Abstract
Estrogen receptor α (ERα) is a crucial transcriptional regulator in breast cancer, but estrogens mediate their effects through two estrogen receptors, ERα and ERβ, subtypes that have contrasting regulatory actions on gene expression and the survival and growth of breast cancer cells. Here, we examine the impact of ERβ on the ERα-p53 loop in breast cancer. We found that ERβ attenuates ERα-induced cell proliferation, increases apoptosis, and reverses transcriptional activation and repression by ERα. Further, ERβ physically interacts with p53, reduces ERα-p53 binding, and antagonizes ERα-p53-mediated transcriptional regulation. ERα directs SUV39H1/H2 and histone H3 lys9 trimethylation (H3K9me3) heterochromatin assembly at estrogen-repressed genes to silence p53-activated transcription. The copresence of ERβ in ERα-positive cells abrogates the H3K9me3 repressive heterochromatin conformation by downregulating SUV39H1 and SUV39H2, thereby releasing the ERα-induced transcriptional block. Furthermore, the presence of ERβ stimulates accumulation of histone H3 lys4 trimethylation (H3K4me3) and RNA polymerase II (RNA Pol II) on ERα-repressed genes, inducing H3K4me3-associated epigenetic activation of the transcription of these repressed genes that can promote p53-based tumor suppression. ERβ also reduced corepressor N-CoR and SMRT recruitment by ERα that could attenuate the crosstalk between ERα and p53. Overall, our data reveal a novel mechanism for ERβ’s anti-proliferative and pro-apoptotic effects in breast cancer cells involving p53 and epigenetic changes in histone methylation that underlie gene regulation of these cellular activities.
Similar content being viewed by others
Introduction
Estrogens mediate their effects on gene regulation and cell proliferation through two estrogen receptors, ERα and ERβ, which belong to the superfamily of nuclear receptors and function as ligand-regulated transcription factors [1]. About 70% of human breast cancers express ERα [2], and the majority of ERα-positive breast cancers also express ERβ [3,4,5]. ERα and ERβ share 96% amino acid identity in their DNA-binding domains and 53% identity in their carboxy-terminal ligand-binding domains, but only 20% identity in their transcription activation function-1 (AF-1) region [6], suggesting that the two ERs might have distinct as well as overlapping roles in regulating gene transcription activities. In addition, because ERα and ERβ are able to heterodimerize [7], they could have joint as well as separate, comodulatory actions in breast cancer. Transcriptomic analyses delineating the impact of ERβ on gene networks have revealed that ERβ is generally a negative regulator of ERα [7,8,9,10,11,12].
The tumor suppressor TP53 (p53), like ERα, is another critical transcription factor that regulates many downstream targets responsible for control of the cell cycle and apoptosis, and it is the most commonly mutated gene in human cancers, including breast cancer. p53 normally induces G1 arrest by transcriptionally activating the cyclin-dependent kinase inhibitor p21 (CDKN1A) [6], and numerous strategies to reactivate or restore mutated p53 function to suppress tumor growth are being explored [13,14,15,16]. It was reported that if p53 is functionally incapacitated in ER-positive breast cancer, ERα could repress p53-mediated transcriptional activation and prevent p53-dependent apoptosis [17,18,19]. Disruption of this ERα-p53 interaction and the restoration of p53 function, which could promote p53-based tumor suppression, might be an appealing strategy for anticancer therapy in breast tumors.
Recent studies have identified a p53-ERα loop in breast cancer involving extensive crosstalk and mutual regulation of their activities [18, 20]. ERα antagonizes the pro-apoptotic function of p53, promoting cancer cell survival [17]. Clinical findings indicate that the presence of wild-type p53 correlates with positive therapeutic response to the endocrine agent tamoxifen [18], whereas high p53 accumulation in breast tumors is a strong predictor of both early and late recurrence in postmenopausal ERα-positive breast cancer patients treated with aromatase inhibitors [21]. Despite studies on interrelationships between ERα and p53 in breast cancer, very little is known about the cellular mechanisms by which ERβ might be impacting p53 function or the crosstalk and interrelationships between ERα and p53. Therefore, to explore these, we examined the effect of ERβ on the ERα-p53 loop in breast cancer cells. In addition, because chromatin conformation is crucial in the control of gene transcription, and multiple modifications of histone tails play essential roles in the control of chromatin organization and gene transcription [22], we examined the effect of ERβ on these chromatin modifications and alterations in target gene regulations.
Materials and Methods
Cell Culture, Adenovirus Infection and siRNA Transfection of Cells
MCF-7 cells (from ATCC) were cultured in Minimal Essential Medium (Sigma) supplemented with 10% calf serum (HyClone), 100 μg/mL penicillin-streptomycin (Invitrogen), and 25 μg/mL gentamicin (Invitrogen). MDA-MB-157 cells were obtained from ATCC and cultured in Leibovitz’s L-15 medium (ATCC) supplemented with 10% fetal bovine serum (HyClone) and 100 μg/mL penicillin-streptomycin (Invitrogen). Recombinant adenoviruses were constructed and prepared as described previously [7]. Cells were infected with adenovirus expressing β-galactosidase (AdGal) or ERβ (AdERβ) as described [7, 10]. The short interfering RNA (siRNA)-mediated gene knockdowns (KD) were performed as described [23]. ERα siRNA was purchased from Dharmacon. The siERα sequences are UCAUCGCAUUCCUUGCAAAdTdT and UUUGCAAGGAAUGCGAUGAdTdT. p53 siRNA was purchased from Santa Cruz (sc-29435).
Cell Proliferation, Colony Formation, and Apoptosis Assays
Cell proliferation was monitored using the WST-1 assay (Roche) as described previously [10]. MCF-7 cells were seeded in 24-well plates at a density of 10,000 cells per well and cultured at 37 °C with 5% CO2. The next day, cells were infected with AdGal and AdERβ for 4 h and then treated with 0.1% EtOH (Veh) or 10 nM E2 on day 0 and on days 3 and 6. Cell growth curves monitored the OD450nm values and each point represents the mean ± SEM. Each sample was done in triplicate. Colony formation assays were performed as described [24]. Apoptosis was measured based on DNA content and analyzed by flow cytometry as described previously [24].
Chromatin Immunoprecipitation Assays
ChIP assays were performed as previously described [10]. Experiments were conducted in hormone-depleted MCF-7 cells treated with 0.1% control EtOH vehicle or 10 nM E2 for 45 min. Chromatin was cross-linked with 1% formaldehyde for 10 min at room temperature. Cells were washed with PBS and collected using lysis buffer supplemented with 1× complete protease inhibitor mix (Roche). Chromatin was sonicated in lysis buffer to 300–500 bp size fragments. Antibodies were incubated with cell lysates overnight for chromatin collection and then with Dynabead Protein A and G (Life Technologies) for 6 h. ChIP DNA was isolated using PCR purification kit (Qiagen) per the manufacturer’s instructions and used for quantitative real-time PCR. Amounts of ChIP DNA were normalized to inputs. Antibodies used for ChIP experiments were H3K4me3 (ab8895, Abcam); H3K9me3 (ab8898, Abcam); p53 (DO-1, sc-29435); RNA Polymerase II (29634A, CTD4H8 clone, Upstate); ERα (HC-20, Santa Cruz); ERβ antibodies were a combination of CWK-F12 (produced in our lab) [25], GTX70182 (GeneTex), GR40 (Calbiochem), and PA1-311 (Affinity Bioreagents); N-CoR (sc-1609, Santa Cruz); and SMRT (sc-1610, Santa Cruz).
RNA Isolation and Real-Time PCR
Total RNA was isolated from cells using TRIzol (Invitrogen) according to the manufacturer’s instructions. RNA samples were reverse transcribed using SuperScript II reverse transcriptase (Invitrogen). Quantitative real-time PCR (qRT-PCR) analyses were performed on the ABI Prism 7900HT using SYBR Green PCR Master Mix (Roche). All experiments were repeated at least three times. The expression of target genes was normalized to the reference gene, usually 36B4.
Gene Expression and Gene Ontology Category Analysis
Gene ontology analysis was performed by using the web-based DAVID Bioinformatics Resources database [26, 27]. Hierarchical clustering of data was performed and displayed using Cluster 3.0 and Java TreeView. Conservation of the binding sites was determined using the web-based Cistrome/Galaxy platform (http://cistrome.org/ap).
Immunoprecipitation and Immunoblotting Analysis
Cells were collected using lysis buffer supplemented with 1× complete protease inhibitor mix (Roche). The protein concentrations of the lysates were quantified using a BCA protein assay kit (Pierce). Cell lysates were incubated with primary antibody and Dynabead protein A and G (Life Technologies) overnight on a rotator. Immunoblotting analyses were performed as previously described [28]. Proteins were separated on SDS-PAGE gels and transferred to nitrocellulose membranes. Antibodies used for immunoprecipitation and immunoblotting were p53 antibody (DO-1, sc-29435); ERα antibody (F10, Santa Cruz); ERβ antibodies (CWK-F12 produced in our lab [25, 29]; H3K4me3 (ab8895, Abcam); H3K9me3 (ab8898, Abcam); N-CoR (sc-1609, Santa Cruz); and SMRT (sc-1610, Santa Cruz).
Breast Cancer Patient Survival Analysis
The survival analysis was conducted as described in [10]. The lists were interrogated using Oncomine concepts analysis [30]. We assessed the clinical relevance of these genes in patients with breast cancer using Oncomine datasets. The breast cancer patients were stratified based on the average expression value of the genes. The top 30% and bottom 30% of patients were used as high- or low-expressing groups for the computation of Kaplan–Meier curves. Survival curves were drawn and compared with Cox–Mantel log-rank test and Gehan–Breslow–Wilcoxon tests using GraphPad Prism. The log2 median-centered intensity expression values for genes were obtained from the Oncomine database.
Statistical Analysis
Data were analyzed using one-way analysis of variance (ANOVA) with Bonferroni post hoc test (GraphPad, San Diego, CA, USA). Results are the means ± SEM of ≥3 independent experiments. The threshold value P < 0.05 was considered to be statistically significant.
Results
ERβ Suppresses Estradiol-Stimulated Proliferation and Anti-Apoptotic Activity in ERα-Positive Breast Cancer Cells
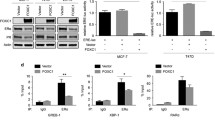
It is well documented that estrogens stimulate cell proliferation in ERα-positive breast cancer cells [31]. To understand the role of ERβ in regulating breast cancer cell growth, we used adenovirus-mediated gene delivery of ERβ into MCF-7 cells, and we validated ERβ expression at the protein level by western blot (Fig. 1a). As shown in Fig. 1b, the copresence of ERβ in ERα breast cancer cells greatly reduced cell proliferation stimulated by E2 (Fig. 1b). We also assessed the ability of cells to form colonies in soft agar. ERα-containing cells generated a large number of colonies with E2 treatment, and the copresence of ERβ markedly reduced the number of colonies formed (Fig. 1c).
ERβ attenuates estradiol-stimulated cell proliferation and colony formation and increases apoptosis. a Western blots show the expression of ERβ in MCF-7 cells infected with Ad-ERβ and treated with 0.1% EtOH (Veh) or 10 nM E2. β-actin was used as loading control. b ERβ attenuates cell proliferation stimulated by E2. MCF-7 cells were seeded in 24-well plates and treated with 0.1% EtOH (Veh) or 10 nM E2 for 6 days. Cell proliferation was measured using the WST-1 assay. *P < 0.05; **P < 0.01. c E2 increases colony formation in ERα-containing cells and ERβ copresence with ERα reduces colony formation with E2 treatment. d ERβ increases apoptosis of MCF-7 cells. *P < 0.05; **P < 0.01
Prior studies have suggested that ERα suppresses p53-dependent apoptosis in breast cancer [17]. To assess the effect of ERβ on cell apoptosis, we performed flow cytometry analyses. As shown in Fig. 1d, E2 reduced apoptosis in cells containing ERα, as expected, but most notable was that expression of ERβ greatly increased the percent of cells undergoing apoptosis (from 4 to 16%) and E2 no longer affected this high level of cell apoptosis (Fig. 1d).
ERβ Antagonizes ERα-Mediated Transcriptional Repression
To investigate the molecular mechanisms involved in the ERβ-mediated anti-proliferative and pro-apoptotic effects in ERα-positive breast cancer cells, we performed RNA-seq to profile the alterations of gene expression in ERα cells and ERα+ERβ cells in response to E2 treatment [8]. We analyzed the transcriptome between the E2-treated and control vehicle samples in ERα cells (ERα cells with E2 treatment vs ERα cells with Veh treatment, fold change (FC) ≥ 2) and identified the genes significantly up- or down-regulated by E2: 926 genes were up-regulated and 1288 genes were down-regulated (Fig. 2a). Within the set of estrogen-regulated genes, we also compared the expression of genes that were significantly up- or down-modulated by ERβ (ERα+ERβ cells with E2 treatment vs ERα cells with E2 treatment, FC ≥ 2) and found that 125 (13%) of these ERα up-regulated genes were down-regulated by ERβ and 674 (52%) of these ERα down-regulated genes were up-regulated by ERβ (Fig. 2a). In essence, the suppression of more than half of the ERα-repressed genes was reversed in the presence of ERβ.
ERβ antagonizes ERα-mediated transcriptional repression. a Hierarchical clustering shows the expression profile for E2-regulated genes (10 nM E2 for 24 h). Cluster I, E2-repressed genes up-regulated by ERβ (674 genes). Cluster II, E2-activated genes down-regulated by ERβ (125 genes). The RNA-Seq dataset accession number is GSE56066. b Comparison of the cluster I genes possessing ERα and ERβ binding sites. The ChIP-Seq dataset accession number is GSE42348. c Gene ontology enrichment analysis of the cluster I genes. All data were derived using the DAVID bioinformatics tool. d Patient survival analysis in breast cancer datasets. All the datasets were obtained from the Oncomine database. e Comparison of average expression levels of genes based on survival data of patients from the Oncomine database
To determine whether the 674 genes repressed by ERα but activated by ERβ were directly influenced by ERα and ERβ, we compared ERα and ERβ genomic occupancy with the 674 genes in MCF-7 cells. By chromatin immunoprecipitation followed by sequencing (ChIP-Seq) data analysis [10], we found that there were 289 genes with ERα binding sites in ERα cells and 221 genes with ERβ binding sites in ERα+ERβ cells within 50 kb of the transcription start site (TSS). Of these genes, 172 had both ERα and ERβ binding sites (Fig. 2b). Gene ontology (GO) analysis revealed that these 172 genes were mainly associated with secretion, cell adhesion, and cell signaling (Fig. 2c).
To explore the clinical relevance of these 172 genes in patients with breast cancer, we used the Oncomine database to analyze the expression of these genes in three large clinical datasets and observed that high expression of these genes was significantly associated with better prognosis in breast cancer patients. In these clinical studies [32,33,34], patients with tumors having a high level of expression of these genes had better overall survival than those with tumors expressing low levels of these genes (Fig. 2d, e).
ERβ Physically Interacts with p53 and Regulates a Set of Common Genes
The transcription factor p53 is crucial in regulating the cell cycle, apoptosis, senescence, and genome stability. Because we found that many of the ERβ-regulated genes were also classical p53 target genes, we wondered whether ERβ might be interacting with p53. By coimmunoprecipitation analyses of extracts from MCF-7 cells coexpressing ERβ, we found that ERβ physically interacted with p53 (Fig. 3a). Interactions between ERβ and endogenous p53 in these extracts could also be detected in reciprocal coimmunoprecipitations.
ERβ and p53 regulate a common set of gene targets. a ERβ interacts with p53. MCF-7 cells were infected with Ad-ERβ. Whole cell lysates were immunoprecipitated using antibody against the indicated protein. Immunocomplexes were then immunoblotted using antibodies against ERβ or p53. b Comparison of genes regulated by ERβ and by p53 in MCF-7 cells. The RNA-Seq dataset accession numbers are GSE56066 and GSE47042. c Hierarchical clustering of the 782 genes commonly regulated by ERβ and p53. d Gene ontology enrichment analysis of the genes commonly up-regulated (318 genes) or down-regulated (377 genes) by both ERβ and p53. e Comparison of commonly regulated genes possessing both ERβ and p53 binding sites. The ERβ ChIP-Seq dataset accession number is GSE42348. The p53 ChIP-Seq dataset is from [35]
Nutlin-3a is a small molecule that activates the p53 pathway by disrupting the p53-MDM2 interaction. Activation of p53 by nutlin-3a induces apoptosis, cell cycle arrest, and growth suppression in cancer cells [15]. We next compared the expression of genes regulated by ERβ [8] and compared them with the genes stimulated by Nutlin-3a treatment of MCF-7 cells [36]. By analyzing RNA-Seq data, we found that 5212 genes were significantly up- or down- regulated by ERβ (ERα+ERβ cells with E2 treatment vs ERα cells with E2 treatment, FC ≥ 2), and 1702 genes were significantly up- or down-regulated after Nutlin-3a treatment (Nutlin-3a vs Veh, FC ≥ 2). Among these genes, 782 were commonly regulated by both ERβ and p53 (Fig. 3b). Hierarchical clustering of the set of genes commonly regulated by both ERβ and p53 revealed two major clusters: genes that were commonly up-regulated by ERβ and p53 (318 genes) and genes that were commonly down-regulated by ERβ and p53 (377 genes) (Fig. 3c). Within the common set of 782 genes regulated by both ERβ and p53, about 84% of the ERβ up-regulated genes were also up-regulated by p53, and almost 93% of the ERβ down-regulated genes were also down-regulated by p53, so there is nearly a complete overlap of ERβ- and p53-regulated genes in these cells.
GO analysis revealed that the genes commonly up-regulated by ERβ and p53 were mainly associated with cell death and apoptosis, and the genes that were commonly down-regulated by ERβ and p53 were primarily associated with cell cycle processes (Fig. 3d). Overall, this analysis revealed an activating role for ERβ and p53 in the control of genes regulating cell death and apoptosis, and a repressive role in the processes associated with cell cycle and mitosis. In essence, ERβ can replicate many of the gene regulatory effects of activated p53, and like p53 can reverse many of the gene actions of ERα that drive proliferation of breast cancer cells.
To determine whether the genes commonly regulated by both ERβ and p53 were directly influenced by ERβ and p53, we analyzed ChIP-Seq data (10,36) and compared the ERβ and p53 genomic occupancy with the co-up and co-down regulated genes in MCF-7 cells. Among the 318 genes commonly up-regulated by both ERβ and p53, there were 157 genes with ERβ binding sites and 172 genes with p53 binding sites within 50 kb of the TSS (Fig. 3e), and of these, 104 had both ERβ and p53 binding sites. Among the 377 down-regulated genes, there were 172 genes with ERβ binding sites, 188 genes with p53 binding sites, and 108 with both ERβ and p53 binding sites (Fig. 3e).
ERβ Activates ERα-Repressed Genes in a p53-Dependent Manner
The fact that ERβ physically interacts with p53 and regulates a set of common genes with p53 raised the possibility that the activation by ERβ of ERα-repressed genes might be dependent on p53. To determine whether ERβ-mediated transcriptional activation of ERα-repressed genes is p53-dependent, we examined the expression of these genes in the ERα+ERβ cells depleted of p53 by transfection with p53 siRNA. Endogenous p53 knockdown was confirmed at the messenger RNA (mRNA) and protein levels by RT-PCR and Western blot immunoassay (Fig. S1, panels A and B). As shown in Fig. 4a, p53 depletion severely impaired the ERβ activation of ERα-repressed genes. The mRNA levels of p21, BTG2, MDM2, and DR5 were significantly decreased in ERα+ERβ cells transfected with p53 siRNA. We also examined the ERβ-mediated transcriptional activation in the p53-null breast cancer cell line, MDA-MB-157 (MDA-MB-157 p53 −/− line) (Fig. S1, panels C and D). In these MDA-MB-157 cells in which we expressed ERβ using adenovirus-mediated gene delivery, little or no increase of these repressed genes was observed (Fig. 4b). These results suggest that p53 is required for ERβ-induced transcriptional activation of ERα-repressed genes.
ERβ activates ERα-repressed genes in a p53-dependent manner. a qRT-PCR analysis of estrogen-repressed genes in control and p53-depleted MCF-7 cells. MCF-7 cells were transfected with p53 siRNA for 72 h. b qRT-PCR analysis of estrogen-repressed genes in wild-type MCF-7 cells (p53+/+) and MDA-MB-157 (p53−/−)cells. c The mRNA and protein levels of p53 in MCF-7 ERα and ERα+ERβ cells. d p53 enhances the activation by ERβ of ERα-repressed genes. MCF-7 cells were treated with the p53-activating agent Nutlin-3a (10 μM) for 24 h; total RNA was extracted and the mRNA levels of the indicated genes were analyzed by qRT-PCR. *P < 0.05; **P < 0.01
To assess whether ERβ affects the expression of p53 and then regulates the p53 pathway, we examined the mRNA and protein levels of p53 in ERα cells and ERα+ERβ cells. As shown in Fig. 4c, the presence of ERβ did not change the level of p53 mRNA or protein. However, treatment of ERα+ERβ cells with the p53 pathway activator Nutlin-3a enhanced the activation of ERα-p53-repressed genes (Fig. 4d). The mRNA levels of p21, BTG2, MDM2, and DR5 were all significantly elevated in ERα+ERβ cells treated with Nutlin-3a (Fig. 4d). Thus, the effect that ERβ has on ERα-repressed genes could be enhanced by active p53.
ERβ Competes with ERα for p53-Mediated Transcriptional Activation
Prior studies have shown that ERα suppresses p53-mediated transcriptional activation and p53-dependent apoptosis in breast cancer [17, 18]. Given that we found that ERβ antagonizes ERα-mediated transcriptional repression and ERβ also can form a complex with p53, we investigated whether ERβ has an effect on p53-mediated transcriptional activation. To test this, we examined the effect of ERβ on the transcription of p53 direct target genes, i.e., the proliferation associated genes p21, MDM2, BTG2, ATF3, and GDF15, and the apoptosis associated genes DR5, Bax1, and TRAF4. Consistent with previous studies, ERα inhibited p53 transcriptional activities, and we observed the mRNA level of p53 target genes to be decreased in MCF-7 cells upon E2 treatment (Fig. 5a). Moreover, this E2-mediated repression was ERα dependent, as siRNA knockdown of ERα resulted in an increase in the RNA level of p53 target genes (Fig. S2). By contrast, ERβ significantly increased the mRNA levels of p53 target genes in the MCF-7 cells (Fig. 5a). These observations suggest that ERα represses p53-mediated transcriptional activities, whereas ERβ activates p53-mediated transcriptional activities.
ERβ antagonizes ERα-p53-mediated transcriptional repression. a qRT-PCR analysis of p53 target genes in ERα cells and ERα+ERβ cells with control vehicle and 10 nM E2 treatment for 24 h. b Interaction of ERα-p53 and ERβ-p53 in MCF-7 cells. Cell lysates were obtained from cells and immunoprecipitated with p53 antibody. Immunocomplexes were then immunoblotted using antibodies against the indicated protein (ERα, ERβ, p53). c ChIP-Seq tracks the occupancy of ERα and ERβ at the p21 locus in ERα cells and ERα+ERβ cells. The ChIP-Seq dataset accession number is GSE42348. d ChIP-qPCR confirms the recruitment of ERα and ERβ to the p21 locus in ERα cells and ERα+ERβ cells. *P < 0.05; **P < 0.01
To assess the interaction of p53 with ERα and ERβ, we performed coimmunoprecipitation experiments using p53 antibody followed by immunoblotting with ERα and ERβ antibodies. Coimmunoprecipitation experiments revealed an interaction of p53-ERα and p53-ERβ (Fig. 5b). In ERα cells, p53 coimmunoprecipitated with ERα, and the presence of ERβ reduced the interaction between p53 and ERα (anti-p53 IP ERα immunoblot lanes 3 and 4 are less intense than lanes 1 and 2). Furthermore, the interaction between p53 and ERβ was observed in cells containing both ERα and ERβ (ERβ immunoblot lanes 3 and 4 are apparent) (Fig. 5b).
To confirm that ERβ competes preferentially with ERα for p53-mediated gene transcriptional regulation, we mapped the genomic localization of ERα and ERβ using ChIP-Seq [10] and identified the occupancy of ERα and ERβ on the p53 target gene, p21. In ERα cells, ERα was recruited to the p21 gene locus, and with the copresence of ERβ, ERβ abrogated the recruitment of ERα to the p21 gene locus. Instead, ERβ was recruited to the same binding sites on the p21 gene (Fig. 5c). We also performed chromatin immunoprecipitation followed by quantitative PCR (ChIP-qPCR), and in agreement with the ChIP-Seq analysis, the ChIP-qPCR results also showed that ERα was recruited to the p21 gene promoter locus in ERα cells and that the presence of ERβ suppressed the recruitment of ERα and increased recruitment of ERβ to the p21 gene locus (Fig. 5d). These results further confirm that ERβ competes with ERα for occupancy of this estrogen-repressed gene and that these receptors have opposite effects on p53-mediated transcriptional activity.
ERβ Abrogates H3K9me3-Mediated Heterochromatin Silencing of ERα-Repressed Genes
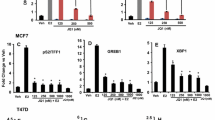
Chromatin conformation plays a critical role in regulating gene transcription and gene silencing. Previous studies have shown that the H3K9me3 repressive histone conformation is enriched on the promoters of both p53 anti-proliferation and pro-apoptotic targets [37]. To investigate whether ERβ affects chromatin-modifying enzymes that could alter chromatin accessibility and allow ERα-repressed genes to become activated in the presence of ERβ, we examined the expression of histone modification enzymes and identified two histone methyltransferases, SUV39H1 and SUV39H2, which specifically tri-methylate histone H3 at lys 9 (H3K9me3). Notably, RNA-Seq data analysis and qPCR results both showed that estradiol activated the transcription of SUV39H1 and SUV39H2 in MCF-7 cells, whereas ERβ repressed the expression of these two genes in control vehicle and in E2-treated cells (Fig. 6a).
ERβ induces alteration in chromatin accessibility. a ERβ decreases the mRNA levels of SUV39H1 and SUV39H2 which are stimulated by E2 in ERα cells. b ERβ abrogates H3K9me3-mediated heterochromatin silencing of ERα-repressed genes. ChIP assays were performed using anti-H3K9me3, anti-N-CoR, and anti-SMRT in ERα cells and ERα+ERβ cells. c ERβ potentiates H3K4me3-mediated transcriptional activation of ERα-repressed genes. ChIP assays were performed using anti-H3K4me3 and anti-RNA pol II in ERα cells and ERα+ERβ cells. *P < 0.05; **P < 0.01
SUV39H1 and SUV39H2 are histone code writers responsible for establishing and maintaining the H3K9me3 heterochromatin mark [38]. Previous studies have shown that downregulating SUV39H1 alone was sufficient to overcome the H3K9me3 repressive chromatin conformation barrier [37]. As we observed that the presence of ERβ down-regulates the expression of SUV39H1/H2 and abrogates estradiol-induced transcriptional repression, we investigated the effect of ERβ on the H3K9me3 repressive heterochromatin mark. We examined how alterations in H3K9me3 marks affect the expression of estrogen-repressed genes by analyzing the relative enrichment of H3K9me3 on these genes. Consistent with increased expression of SUV39H1 and SUV39H2 in ERα cells treated with E2, ChIP analysis with anti-H3K9me3 antibody showed that H3K9me3 was significantly increased in ERα cells treated with E2, and that ERβ abrogated the H3K9me3 increase on this ERα-repressed p21 gene (Fig. 6b). These results demonstrate that E2-ERα activates the transcription of SUV39H1 and SUV39H2 and maintains higher levels of the H3K9me3 heterochromatin mark on estrogen-repressed gene loci. However, when ERβ was present, it represses the transcription of SUV39H1 and SUV39H2, thus removing the H3K9me3-repressive chromatin conformation barrier for this ERα-repressed gene.
N-CoR (nuclear receptor corepressor) and SMRT (silencing mediator for retinoid and thyroid hormone receptors) are nuclear receptor co-repressors, both of which are recruited to chromatin by nuclear hormone receptors to regulate gene transcription [18, 39, 40]. Given that ERα was reported to recruit N-CoR and SMRT to repress p53-mediated transcriptional activation [18], we were interested in exploring whether the copresence of ERβ affected the recruitment of nuclear receptor transcriptional repressive complexes on estrogen-repressed genes. We analyzed the occupancy of N-CoR and SMRT on the p21 gene in ERα cells and ERα+ERβ cells. ChIP experiments documented that ERβ markedly decreased occupancy of N-CoR and SMRT on the p21 gene in the presence and absence of E2 (Fig. 6 b). ERβ also eliminated the increased recruitment of H3K9me3 to p21 by E2 and ERα (Fig. 6b).
ERβ Potentiates H3K4me3-Mediated Epigenetic Activation of Estrogen-Repressed Genes
Chromatin remodeling could modulate transcription by blocking or facilitating transcription factor access to DNA. H3K4me3 is a well-known marker that is correlated with gene activation, and previous studies showed that H3K4me3 enhances p53-dependent transcription [41]. Given that we observed that ERβ could remove the H3K9me3 repressive heterochromatin barrier for estrogen-repressed genes and increase the levels of histone active H3K4me3 marks, we analyzed the enrichment of H3K4me3 on the ERα-repressed genes in ERα cells and in ERα+ERβ cells. ChIP analysis with anti-H3K4me3 antibody showed that the occupancy of H3K4me3 on the ERα-repressed gene p21 was markedly increased in MCF-7 cells in the copresence of ERβ (Fig. 6c). We also investigated the recruitment of RNA Pol II on ERα-repressed genes in ERα and in ERα+ERβ cells. ChIP analysis using anti-RNA Pol II showed that the accumulation of RNA Pol II at the p21 gene was greatly increased (by fivefold) in the ERα+ERβ cells (Fig. 6c), supporting that increased RNA Pol II recruitment likely contributes to the activation of ERα-repressed genes in cells containing ERβ.
Discussion
Our studies have delineated a modulatory role for ERβ on the p53-ERα loop in breast cancer that involves crosstalk and mutual regulation of the activities of these transcription factors [18, 20]. ERα antagonizes the pro-apoptotic function of p53, promoting cancer cell survival [17], but very little was known about how ERβ might impact p53 function, and the crosstalk and interrelationships between ERα and p53. To understand the molecular mechanisms responsible for the anti-proliferation and pro-apoptosis roles of ERβ we observed, we analyzed the genes regulated by estradiol and by ERα and ERβ and found that 52% of estradiol down-regulated genes in cells containing only ERα were activated by ERβ and 13% of estradiol up-regulated genes by ERα were repressed by ERβ. Of note, more than half of the ERα-repressed genes had their expression increased by ERβ. While it is well known that ERα can both stimulate and suppress gene transcription [42,43,44,45,46], previous studies have focused mainly on the genes that are up-regulated following estrogen stimulation, whereas the molecular mechanisms that lead to gene repression are known to a much more limited extent [40, 46]. Notably, more than half of the ERα-repressed genes were activated by ERβ, suggesting that the reactivation of ERα-repressed genes by ERβ may be an important aspect underlying its attenuation of E2-ERα-induced cell proliferation and anti-apoptosis.
In our studies, ERβ acted as a growth suppressor, exerting anti-proliferative and pro-apoptotic effects in ERα-positive breast cancer cells. Previous studies have shown that ERβ induced the expression of GADD45A, BTG2, and PUMA [47, 48], which provided clues to the potential for crosstalk between p53 and ERβ signaling pathways. We found that ERβ physically interacted with p53 and that nearly half of the p53-regulated genes were also regulated by ERβ. Furthermore, both ERβ and p53 were found to play an activating role in the control of genes regulating cell death and apoptosis, and a repressive role in processes associated with cell cycle progression. ERβ did not change the expression of p53 at either the mRNA or protein levels, but ERβ activated ERα-repressed genes in a p53-dependent manner, supporting the notion that ERβ is a novel activator of the p53 pathway and can act as a tumor suppressor in breast cancer cells. Notably, ERβ had a modulatory effect even in the absence of ligand, consonant with the known substantial ligand-independent activity of ERβ [10].
Transcriptional regulation by DNA-binding transcription factors such as ERα, ERβ, and p53 is a multi-step process that involves numerous protein complexes, the basal transcription machinery, and the remodeling of chromatin structure ( [22, 49,50,51]). Genome-wide analyses of chromatin modifications have indicated that H3K9me3 is enriched in heterochromatin, which is in a transcriptionally repressed state generally linked to gene silencing [52], whereas H3K4me3 is enriched around the transcriptional start site of active promoters where TFIID and RNA polymerase are also present [53, 54]. SUV39H1 and SUV39H2 are histone methyltransferases that specifically trimethylate histone H3 at lys 9 to mark the histone H3 for a “closed” chromatin conformation [55]. H3K9me3 repressive histone conformation is enriched on the promoters of both p53 anti-proliferation and pro-apoptotic target genes, and SUV39H1 silencing alone is sufficient to reduce the levels of H3K9me3 on the promoters of p53 anti-proliferation and pro-apoptotic targets and overcome this repressive chromatin conformation barrier [37]. In our study, we observed that ERβ attenuation of estrogen-induced cell proliferation occurred by preventing ERα-p53-mediated transcriptional repression. ERα directed SUV39H1/H2 mediated H3K9me3 heterochromatin assembly at estrogen-repressed genes upon 17β-estradiol (E2) exposure, and ERβ abrogated this H3K9me3 repressive chromatin conformation by downregulating SUV39H1/H2 and inducing H3K4me3-mediated epigenetic activation of estrogen-repressed genes.
p53 plays a crucial role in various physiological processes, including apoptosis, cell cycle progression, and genome stability [56]. More than 50% of human cancers contain p53 that is inactivated by mutations or deletion, and reactivation or restoration of functional p53 to suppress tumors has been suggested as an appealing strategy for anticancer therapy. In breast cancers, p53 is only mutated in approximately 20–30% of cases [57], but despite harboring wild-type p53, ERα could recruit nuclear receptor corepressor (N-CoR) to repress p53-mediated transcriptional activation and prevent p53-dependent apoptosis [17,18,19], thereby functionally incapacitating p53’s tumor-suppressive actions. Of interest, we observed that the copresence of ERβ reduced N-CoR and SMRT recruitment by ERα, which could attenuate the crosstalk between ERα and p53.
Hormonal therapy together with radiation therapy can improve local tumor control, decrease distant metastases and improve survival of patients with breast cancer [58, 59]. One of the proposed explanations is that radiation disrupts the ERα-p53 interaction leading to restoration of p53 function. In our study, we found that ERβ disrupted ERα-p53 interaction, antagonized ERα-p53-mediated transcriptional repression, and reactivated p53-mediated gene transcriptional activity. In addition, introduction of ERβ together with activation of the p53 pathway using Nutlin-3a resulted in an additive activation of ERα-repressed genes, which could promote tumor suppression.
Our studies examined the effects of ERβ on the crosstalk between ERα and p53, and we therefore studied the effects with both ER subtypes present. However, the actions of ERβ may well show context- and cell type-dependent differences. In this regard, ERβ has been reported recently to be present in the majority of breast cancer stem cells, with ERβ expression declining as stem cells differentiate [60]. In breast cancer, stem cells that lack ERα but contain ERβ, E2, or the ERβ agonist DPN increased mammosphere formation and proliferation, and changed the breast stem cell metabolism. Further, an ERβ antagonist, PHTPP, when combined with tamoxifen, reduced tumor growth over that achieved by tamoxifen alone [60]. Hence, ERβ may function as a proliferative signal in some contexts when ERα is absent [60].
We have also shown that ERβ in the absence of ERα can alter breast cancer cell metabolism [10]. ERβ greatly increased the genomic binding and expression level of RIP 140, a nuclear receptor coregulator and critical regulator of metabolism, resulting in increased expression of adipogenesis genes and elevated triglyceride levels in breast cancer cells expressing ERβ in the absence of ERα. In addition, ERα and ERβ were found to select distinct, only partially overlapping chromatin binding sites, with binding site selection and cistrome redistribution being profoundly altered when both receptors were present [10]. Hence, the role of ERβ is no doubt context- and cell type-dependent and appears to differ when ERα is copresent or not.
Chromatin conformation is crucial in regulating gene transcription by blocking or facilitating the access of transcription factors to DNA. Previous studies have shown that p53 anti-proliferation and pro-apoptotic target promoters are enriched in H3K9me3 repressive heterochromatin marks in unstressed cells [37]. SUV39H1 downregulation was alone sufficient to lead to a switch from the H3K9me3-associated repressive to the H3K4me3-associated active transcription state of p53 anti-proliferation and pro-apoptotic target promoters [37]. In this study, we found that estradiol could induce the expression of SUV39H1 and SUV39H2 and direct SUV39H1/H2 H3K9me3 heterochromatin assembly at ERα-repressed genes to silence transcription. Introduction of ERβ down-regulated SUV39H1 and SUV39H2 and released the H3K9me3-mediated heterochromatin silencing of ERα-repressed genes. In addition, the presence of ERβ induced the accumulation of H3K4me3 and RNA Pol II on ERα-repressed genes, with epigenetic activation of the transcription of p21, the ERα-repressed and p53-stimulated gene. Taken together, our data reveal a novel mechanism by which ERβ affects the p53-ERα loop in breast cancer and overall reinforces anti-proliferative and pro-apoptotic activities. These interrelationships should affect not only breast cancer growth and progression but also the responsiveness of ER-containing breast cancers to endocrine and other cancer therapies.
Abbreviations
- ChIP:
-
Chromatin immunoprecipitation
- ERα:
-
Estrogen receptor alpha
- ERβ:
-
Estrogen receptor beta
- E2:
-
17β-estradiol
References
Pettersson K, Gustafsson JA (2001) Role of estrogen receptor beta in estrogen action. Annu Rev Physiol 63:165–192
Stanford JL, Szklo M, Brinton LA (1986) Estrogen receptors and breast cancer. Epidemiol Rev 8:42–59
Katzenellenbogen BS, Frasor J (2004) Therapeutic targeting in the estrogen receptor hormonal pathway. Semin Oncol 31:28–38
Kurebayashi J, Otsuki T, Kunisue H, Tanaka K, Yamamoto S, Sonoo H (2000) Expression levels of estrogen receptor-alpha, estrogen receptor-beta, coactivators, and corepressors in breast cancer. Clin Cancer Res 6:512–518
Saji S, Hirose M, Toi M (2005) Clinical significance of estrogen receptor beta in breast cancer. Cancer Chemother Pharmacol 56(Suppl 1):21–26
el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM et al (1993) WAF1, a potential mediator of p53 tumor suppression. Cell 75:817–825
Chang EC, Frasor J, Komm B, Katzenellenbogen BS (2006) Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology 147:4831–4842
Gong P, Madak-Erdogan Z, Li J, Cheng J, Greenlief CM, Helferich W et al (2014) Transcriptomic analysis identifies gene networks regulated by estrogen receptor alpha (ERalpha) and ERbeta that control distinct effects of different botanical estrogens. Nucl Recept Signal 12:e001
Lindberg MK, Moverare S, Skrtic S, Gao H, Dahlman-Wright K, Gustafsson JA et al (2003) Estrogen receptor (ER)-beta reduces ERalpha-regulated gene transcription, supporting a “ying yang” relationship between ERalpha and ERbeta in mice. Mol Endocrinol 17:203–208
Madak-Erdogan Z, Charn TH, Jiang Y, Liu ET, Katzenellenbogen JA, Katzenellenbogen BS (2013) Integrative genomics of gene and metabolic regulation by estrogen receptors alpha and beta, and their coregulators. Mol Syst Biol 9:676
Paruthiyil S, Parmar H, Kerekatte V, Cunha GR, Firestone GL, Leitman DC (2004) Estrogen receptor beta inhibits human breast cancer cell proliferation and tumor formation by causing a G2 cell cycle arrest. Cancer Res 64:423–428
Strom A, Hartman J, Foster JS, Kietz S, Wimalasena J, Gustafsson JA (2004) Estrogen receptor beta inhibits 17beta-estradiol-stimulated proliferation of the breast cancer cell line T47D. Proc Natl Acad Sci U S A 101:1566–1571
Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H et al (2006) Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A 103:1888–1893
Shangary S, Wang S (2009) Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol 49:223–241
Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z et al (2004) In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303:844–848
Selivanova G (2010) Therapeutic targeting of p53 by small molecules. Semin Cancer Biol 20:46–56
Bailey ST, Shin H, Westerling T, Liu XS, Brown M (2012) Estrogen receptor prevents p53-dependent apoptosis in breast cancer. Proc Natl Acad Sci U S A 109:18060–18065
Konduri SD, Medisetty R, Liu W, Kaipparettu BA, Srivastava P, Brauch H et al (2010) Mechanisms of estrogen receptor antagonism toward p53 and its implications in breast cancer therapeutic response and stem cell regulation. Proc Natl Acad Sci U S A 107:15081–15086
Liu W, Konduri SD, Bansal S, Nayak BK, Rajasekaran SA, Karuppayil SM et al (2006) Estrogen receptor-alpha binds p53 tumor suppressor protein directly and represses its function. J Biol Chem 281:9837–9840
Berger C, Qian Y, Chen X (2013) The p53-estrogen receptor loop in cancer. Curr Mol Med 13:1229–1240
Yamamoto M, Hosoda M, Nakano K, Jia S, Hatanaka KC, Takakuwa E et al (2014) p53 accumulation is a strong predictor of recurrence in estrogen receptor-positive breast cancer patients treated with aromatase inhibitors. Cancer Sci 105:81–88
Li B, Carey M, Workman JL (2007) The role of chromatin during transcription. Cell 128:707–719
Chang EC, Charn TH, Park SH, Helferich WG, Komm B, Katzenellenbogen JA et al (2008) Estrogen receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol Endocrinol 22:1032–1043
Bergamaschi A, Christensen BL, Katzenellenbogen BS (2011) Reversal of endocrine resistance in breast cancer: interrelationships among 14-3-3zeta, FOXM1, and a gene signature associated with mitosis. Breast Cancer Res 13:R70
Choi I, Ko C, Park-Sarge OK, Nie R, Hess RA, Graves C et al (2001) Human estrogen receptor beta-specific monoclonal antibodies: characterization and use in studies of estrogen receptor beta protein expression in reproductive tissues. Mol Cell Endocrinol 181:139–150
Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC et al (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4:P3
Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Lv WW, Wei HM, Wang DL, Ni JQ, Sun FL (2012) Depletion of histone deacetylase 3 antagonizes PI3K-mediated overgrowth of drosophila organs through the acetylation of histone H4 at lysine 16. J Cell Sci 125:5369–5378
Nelson AW, Groen AJ, Miller JL, Warren AY, Holmes KA, Tarulli GA et al (2017) Comprehensive assessment of estrogen receptor beta antibodies in cancer cell line models and tissue reveals critical limitations in reagent specificity. Mol Cell Endocrinol 440:138–150
Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D et al (2004) ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6:1–6
Katzenellenbogen BS, Kendra KL, Norman MJ, Berthois Y (1987) Proliferation, hormonal responsiveness, and estrogen receptor content of MCF-7 human breast cancer cells grown in the short-term and long-term absence of estrogens. Cancer Res 47:4355–4360
Calza S, Hall P, Auer G, Bjohle J, Klaar S, Kronenwett U et al (2006) Intrinsic molecular signature of breast cancer in a population-based cohort of 412 patients. Breast Cancer Res 8:R34
Kao KJ, Chang KM, Hsu HC (2011) Huang AT correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer 11:143
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW et al (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347:1999–2009
Nikulenkov F, Spinnler C, Li H, Tonelli C, Shi Y, Turunen M et al (2012) Insights into p53 transcriptional function via genome-wide chromatin occupancy and gene expression analysis. Cell Death Differ 19:1992–2002
Janky R, Verfaillie A, Imrichova H, Van de Sande B, Standaert L, Christiaens V et al (2014) iRegulon: from a gene list to a gene regulatory network using large motif and track collections. PLoS Comput Biol 10:e1003731
Mungamuri SK, Benson EK, Wang S, Gu W, Lee SW, Aaronson SA (2012) p53-mediated heterochromatin reorganization regulates its cell fate decisions. Nat Struct Mol Biol 19:478–484 S471
Lehnertz B, Ueda Y, Derijck AA, Braunschweig U, Perez-Burgos L, Kubicek S et al (2003) Suv39h-mediated histone H3 lysine 9 methylation directs DNA methylation to major satellite repeats at pericentric heterochromatin. Curr Biol 13:1192–1200
Perissi V, Jepsen K, Glass CK, Rosenfeld MG (2010) Deconstructing repression: evolving models of co-repressor action. Nat Rev Genet 11:109–123
Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS (2006) Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem 281:16272–16278
Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH et al (2013) H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell 152:1021–1036
Frasor J, Danes JM, Komm B, Chang KC, Lyttle CR, Katzenellenbogen BS (2003) Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562–4574
Green KA, Carroll JS (2007) Oestrogen-receptor-mediated transcription and the influence of co-factors and chromatin state. Nat Rev Cancer 7:713–722
Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M et al (2003) Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115:751–763
Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell 103:843–852
Stossi F, Madak-Erdogan Z, Katzenellenbogen BS (2009) Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol Cell Biol 29:1749–1759
Paruthiyil S, Cvoro A, Tagliaferri M, Cohen I, Shtivelman E, Leitman DC (2011) Estrogen receptor beta causes a G2 cell cycle arrest by inhibiting CDK1 activity through the regulation of cyclin B1, GADD45A, and BTG2. Breast Cancer Res Treat 129:777–784
Dey P, Strom A, Gustafsson JA (2014) Estrogen receptor beta upregulates FOXO3a and causes induction of apoptosis through PUMA in prostate cancer. Oncogene 33:4213–4225
Jenuwein T, Allis CD (2001) Translating the histone code. Science 293:1074–1080
Kouzarides T (2007) Chromatin modifications and their function. Cell 128:693–705
Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ (2007) How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol 14:1025–1040
Peters AH, Mermoud JE, O’Carroll D, Pagani M, Schweizer D, Brockdorff N et al (2002) Histone H3 lysine 9 methylation is an epigenetic imprint of facultative heterochromatin. Nat Genet 30:77–80
Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA (2007) A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88
Czvitkovich S, Sauer S, Peters AH, Deiner E, Wolf A, Laible G et al (2001) Over-expression of the SUV39H1 histone methyltransferase induces altered proliferation and differentiation in transgenic mice. Mech Dev 107:141–153
Vousden KH, Lu X (2002) Live or let die: the cell’s response to p53. Nat Rev Cancer 2:594–604
Olivier M, Langerod A, Carrieri P, Bergh J, Klaar S, Eyfjord J et al (2006) The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res 12:1157–1167
Vinh-Hung V, Verschraegen C (2004) Breast-conserving surgery with or without radiotherapy: pooled-analysis for risks of ipsilateral breast tumor recurrence and mortality. J Natl Cancer Inst 96:115–121
Whelan TJ, Julian J, Wright J, Jadad AR, Levine ML (2000) Does locoregional radiation therapy improve survival in breast cancer? A meta-analysis. J Clin Oncol 18:1220–1229
Ma R, Karthik GM, Lovrot J, Haglund F, Rosin G, Katchy A et al (2017) Estrogen receptor beta as a therapeutic target in breast cancer stem cells. J Natl Cancer Inst 109:1–14
Acknowledgements
This study was funded by a grant from the Breast Cancer Research Foundation (to BSK), and by NIH grant P50AT006268 from the National Center for Complementary and Integrative Health (NCCIH), the Office of Dietary Supplements (ODS), and the National Cancer Institute (NCI) (to BSK). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCCIH, ODS, NCI, or the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 2317 kb)
Rights and permissions
About this article
Cite this article
Lu, W., Katzenellenbogen, B.S. Estrogen Receptor-β Modulation of the ERα-p53 Loop Regulating Gene Expression, Proliferation, and Apoptosis in Breast Cancer. HORM CANC 8, 230–242 (2017). https://doi.org/10.1007/s12672-017-0298-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12672-017-0298-1