Abstract
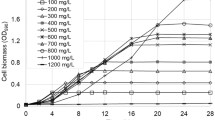
Phenol biodegradation in an aerobic batch reactor was investigated using mixed two co-aggregating strains (Flavobacterium sp. and Acetobacter sp.). Response surface methodology by the Box–Behnken model was used to evaluate the optimal cell growth and phenol degradation conditions. The optimum temperature, pH value and inoculum size were found to be 33 °C, 6.06 and 13 %, respectively. In the conditions, phenol degradation rate and biomass were predicted to be 96.97 % and 410.78 mg/L within the range examined, respectively. Less toxic acetaldehyde, ethanol and acetic ether were identified as main intermediate products from the degraded samples using GC–MS. Substrate inhibition was calculated from experimental biomass growth and phenol degradation parameters using the Haldane equation. Kinetic parameters derived from nonlinear regression with correlation factors (R 2) were 0.9682 for phenol degradation and 0.9594 for biomass growth, respectively. The phenol concentration to avoid substrate inhibition was 278.17 mg/L.







Similar content being viewed by others
References
Agarry SE, Solomon BO, Layokun SK (2008) Kinetics of batch microbial degradation of phenols by indigenous binary mixed culture of Pseudomonas aeruginosa and Pseudomonas fluorescence. Afr J Biotechnol 7:2417–2423
Allsop PJ, Chisti Y, Moo-Young M, Sullivan GR (1993) Dynamics of phenol degradation by Pseudomonas putida. Biotechnol Bioeng 41:572–580
Annadurai G, Juang RS, Lee DJ (2002) Microbiological degradation of phenol using mixed liquors of Pseudomonas putida and activated sludge. Waste Manage 22:703–710
Betancur MJ, Moreno-Andrade I, Moreno JA, Buitrón G, Dochain D (2008) Modeling for the optimal biodegradation of toxic wastewater in a discontinuous reactor. Bioprocess Biosyst Eng 31:307–313
Clesceri LS, Greenberg AE, Eaton AD (1998) Standard methods for the examination of water and wastewater, 20th edn. American Public Health Association, American Water Works Association, Water Pollution Control Federation, Washington DC
Elibol M, Ozer D (2002) Response surface analysis of lipase production by freely suspended Rhizopus arrhizus. Process Biochem 38:367–372
Jing B, Wen JP, Li HM, Jiang Y (2007) Kinetic modeling of growth and biodegradation of phenol and m-cresol using Alcaligenes faecalis. Process Biochem 42:510–517
Kapley A, Tolmare A, Purohit HJ (2001) Role of oxygen in the utilization of phenol by Pseudomonas CF600 in continuous culture. World J Microbiol Biotechnol 17:801–804
Kumar R, Singh R, Kumar N, Bishnoi K, Bishnoi NR (2009) Response surface methodology approach for optimization of biosorption process for removal of Cr(VI), Ni(II) and Zn(II) ions by immobilized bacterial biomass sp. Bacillus brevis Chem Eng J 146:401–407
Léonard D, Lindley ND (1999) Growth of Ralstonia eutropha on inhibitory concentrations of phenol: diminished growth can be attributed to hydrophobic perturbation of phenol hydroxylase activity. Enzyme Microb Tech 25:271–277
Liyana-Parthirana C, Shahidi F (2005) Optimisation of extraction of phenolic compounds from wheat using response surface methodology. Food Chem 93:47–56
Monteiro ÁAMG, Boaventura RAR, Rodrigues AE (2000) Phenol biodegradation by Pseudomonas putida DSM 548 in a batch reactor. Biochem Eng J 6:45–49
Nuhoglu A, Yalcin B (2005) Modeling of phenol removal in a batch reactor. Process Biochem 40:1233–1239
Secula MS, Suditu GD, Poulios I, Cojocaru C, Cretescu I (2008) Response surface optimization of the photocatalytic decolorization of a simulated dyestuff effluent. Chem Eng J 141:18–26
Tsai SC, Tsai LD, Li YK (2005) An isolated Candida albicans TL3 capable of degrading phenol at large concentration. Biosci Biotechnol Biochem 69:2358–2367
Wang JL, Wu WZ, Zhao X (2004) Microbial degradation of quinoline: kinetics study with Burkholderia picekttii. Biomed Environ Sci 17:21–26
Wang Y, Tian Y, Han B, Zhao HB, Bi JN, Cao BL (2007) Biodegradation of phenol by free and immobilized Acinetobacter sp. strain PD12. J Environ Sci 19:222–225
Zeng HY, Jiang H, Xia K, Wang YJ, Huang Y (2010) Characterization of phenol degradation by high-efficiency binary mixed culture. Environ Sci Pollut Res 17:1035–1044
Acknowledgments
This work is supported by Scientific Research Fund of Hunan Provincial Education Department of China through key-project (No. 08A080).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zeng, HY., Cao, XL., Xiong, LB. et al. Microbiological degradation of phenol using two co-aggregating bacterial strains. Environ Earth Sci 71, 1339–1348 (2014). https://doi.org/10.1007/s12665-013-2540-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12665-013-2540-7