Abstract
Background
Nonalcoholic steatohepatitis (NASH), a severe form of nonalcoholic fatty liver disease, can lead to advanced liver damage and has become an increasingly prominent health problem worldwide. Predictive models for early identification of high-risk individuals could help identify preventive and interventional measures. Traditional epidemiological models with limited predictive power are based on statistical analysis. In the current study, a novel machine-learning approach was developed for individual NASH susceptibility prediction using candidate single nucleotide polymorphisms (SNPs).
Methods
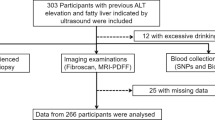
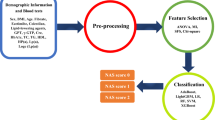
A total of 245 NASH patients and 120 healthy individuals were included in the study. Single nucleotide polymorphism genotypes of candidate genes including two SNPs in the cytochrome P450 family 2 subfamily E member 1 (CYP2E1) gene (rs6413432, rs3813867), two SNPs in the glucokinase regulator (GCKR) gene (rs780094, rs1260326), rs738409 SNP in patatin-like phospholipase domain-containing 3 (PNPLA3), and gender parameters were used to develop models for identifying at-risk individuals. To predict the individual’s susceptibility to NASH, nine different machine-learning models were constructed. These models involved two different feature selections including Chi-square, and support vector machine recursive feature elimination (SVM-RFE) and three classification algorithms including k-nearest neighbor (KNN), multi-layer perceptron (MLP), and random forest (RF). All nine machine-learning models were trained using 80% of both the NASH patients and the healthy controls data. The nine machine-learning models were then tested on 20% of both groups. The model’s performance was compared for model accuracy, precision, sensitivity, and F measure.
Results
Among all nine machine-learning models, the KNN classifier with all features as input showed the highest performance with 86% F measure and 79% accuracy.
Conclusions
Machine learning based on genomic variety may be applicable for estimating an individual’s susceptibility for developing NASH among high-risk groups with a high degree of accuracy, precision, and sensitivity.


Similar content being viewed by others
Data availability
The datasets analyzed during the current study are available in the ZENODO repository and can be accessed from https://doi.org/10.5281/zenodo.4686908.
References
Caligiuri A, Gentilini A, Marra F. Molecular pathogenesis of NASH. Int J Mol Sci. 2016;17:1575.
Adams LA, Feldstein AE. Nonalcoholic steatohepatitis: risk factors and diagnosis. Expert Rev Gastroenterol Hepatol. 2010;4:623–35.
Vespasiani-Gentilucci U, Gallo P, Dell'Unto C, Volpentesta M, Antonelli-Incalzi R, Picardi A. Promoting genetics in non-alcoholic fatty liver disease: combined risk score through polymorphisms and clinical variables. World J Gastroenterol. 2018;24:4835–45.
Vilar-Gomez E, Chalasani N. Non-invasive assessment of non-alcoholic fatty liver disease: clinical prediction rules and blood-based biomarkers. J Hepatol. 2018;68:305–15.
Anstee QM, Seth D, Day CP. Genetic factors that affect risk of alcoholic and nonalcoholic fatty liver disease. Gastroenterology. 2016;150:1728–44.e7.
Kawaguchi T, Shima T, Mizuno M, et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS One. 2018;13:e0185490.
Koo BK, Joo SK, Kim D, et al. Development and validation of a scoring system, based on genetic and clinical factors, to determine risk of steatohepatitis in Asian patients with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol. 2020;18:2592-9.e10.
Gaudillo J, Rodriguez JJR, Nazareno A, et al. Machine learning approach to single nucleotide polymorphism-based asthma prediction. PLoS One. 2019;14:e0225574.
Ostrovski V. New equivalence tests for Hardy–Weinberg equilibrium and multiple alleles. Stats. 2020;3:34–9.
Wang X, Strizich G, Hu Y, Wang T, Kaplan RC, Qi Q. Genetic markers of type 2 diabetes: progress in genome-wide association studies and clinical application for risk prediction. J Diabetes. 2016;8:24–35.
Ma H, Xu CF, Shen Z, Yu CH, Li YM. Application of machine learning techniques for clinical predictive modeling: A cross-sectional study on nonalcoholic fatty liver disease in China. Biomed Res Int. 2018;2018:4304376.
Ho DSW, Schierding W, Wake M, Saffery R, O'Sullivan J. Machine learning SNP based prediction for precision medicine. Front Genet. 2019;10:267.
Yip TC, Ma AJ, Wong VW, et al. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment Pharmacol Ther. 2017;46:447–56.
Canbay A, Kälsch J, Neumann U, et al. Non-invasive assessment of NAFLD as systemic disease-a machine learning perspective. PLoS One. 2019;14:e0214436.
Fialoke S, Malarstig A, Miller MR, Dumitriu A. Application of machine learning methods to predict non-alcoholic steatohepatitis (NASH) in non-alcoholic fatty liver (NAFL) patients. AMIA Annu Symp Proc. 2018;2018:430–9.
Perakakis N, Polyzos SA, Yazdani A, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism. 2019;101:154005.
Chiappini F, Coilly A, Kadar H, et al. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci Rep. 2017;7:46658.
Dai G, Liu P, Li X, Zhou X, He S. Association between PNPLA3 rs738409 polymorphism and nonalcoholic fatty liver disease (NAFLD) susceptibility and severity: A meta-analysis. Medicine (Baltimore). 2019;98:e14324.
Vespasiani-Gentilucci U, Gallo P, Porcari A, et al. The PNPLA3 rs738409 C>G polymorphism is associated with the risk of progression to cirrhosis in NAFLD patients. Scand J Gastroenterol. 2016;51:967–73.
Hotta K, Yoneda M, Hyogo H, et al. Association of the rs738409 polymorphism in PNPLA3 with liver damage and the development of nonalcoholic fatty liver disease. BMC Med Genet. 2010;11:172.
Liu YL, Patman GL, Leathart JB, et al. Carriage of the PNPLA3 rs738409 C>G polymorphism confers an increased risk of non-alcoholic fatty liver disease associated hepatocellular carcinoma. J Hepatol. 2014;61:75–81.
Tan HL, Zain SM, Mohamed R, et al. Association of glucokinase regulatory gene polymorphisms with risk and severity of non-alcoholic fatty liver disease: an interaction study with adiponutrin gene. J Gastroenterol. 2013;49:1056–64.
Ulusoy G, Arinç E, Adali O. Genotype and allele frequencies of polymorphic CYP2E1 in the Turkish population. Arch Toxicol. 2007;81:711–8.
Matsushita N, Hassanein MT, Martinez-Clemente M, et al. Gender difference in NASH susceptibility: roles of hepatocyte Ikkβ and Sult1e1. PLoS One. 2017;12:e0181052.
Noureddin M, Vipani A, Bresee C, et al. NASH leading cause of liver transplant in women: updated analysis of indications for liver transplant and ethnic and gender variances. Am J Gastroenterol. 2018;113:1649–59.
Hashimoto E, Tokushige K. Prevalence, gender, ethnic variations, and prognosis of NASH. J Gastroenterol. 2011;46 Suppl 1:63–9.
Soleymani R, Granger E, Fumera G. F-measure curves: a tool to visualize classifier performance under imbalance. Pattern Recognition. 2020;107146:107146.
Author information
Authors and Affiliations
Contributions
Concept: FG, AAH; design: FG; supervision: AAH, OÖ; materials: AAH; data collection and/or analysis: AAH, FG; literature search: FG; writing: FG, AAH; critical reviews: AAH
Corresponding author
Ethics declarations
Competing interests
FG, AAH and OÖ declare no competing interests.
Ethics statement
The study was performed conforming to the Helsinki declaration of 1975, as revised in 2000 and 2008 concerning human and animal rights, and the authors followed the policy concerning informed consent as shown on Springer.com.
Ethics approval
The ethics committee of Istanbul Gelişim University approved this study (ethical code: 77366270-302.08.01-E.12978, date: 16.11.2020).
Consent to participate
Consent forms were signed by all the participants before being included in the study.
Consent for publication
Not applicable.
Disclaimer
The authors are solely responsible for the data and the contents of the paper. In no way is the honorary editor in chief, editorial board members, the Indian Society of Gastroenterology or the printer/publishers responsible for the results/findings and content of this article.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ghadiri, F., Husseini, A.A. & Öztaş, O. A machine-learning approach for nonalcoholic steatohepatitis susceptibility estimation. Indian J Gastroenterol 41, 475–482 (2022). https://doi.org/10.1007/s12664-022-01263-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12664-022-01263-2