Abstract
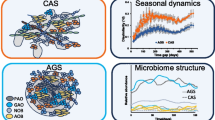
The bacterial and archaeal communities of two full-scale biogas producing plants (P1 and P2), associated with a 999 kW cogeneration unit, both located in North Italy, were analyzed at start up and fully operating phases, by means of various molecular approaches: (i) Automated Ribosomal Intergenic Spacer Analysis; (ii) cloning and sequencing of PCR amplicons of archaeal genes 16Srrna and mcrA; (iii) 16S rDNA high throughput next generation sequencing. P1 and P2 use the same technology and both were fed with cattle manure and corn silage. During the study of P1 also the post digester (fed with pig manure) was analyzed. The aim of this research was to characterize the bacterial and archaeal communities in two very similar plants to profile the core microbiota. The results of this analysis highlighted that the two plants (producing comparable quantities of volatile fatty acids, biogas, and energy) differed in anerobic microbiota (Bacteria and Archaea). Notably the methanogenic community of P1 was dominated by the strict acetoclastic Methanosaeta (Methanothrix) (up to 23.05%) and the unculturable Candidatus Methanofastidiosum (up to 32.70%), while P2 was dominated by the acetoclastic, but more substrate-versatile, Methanosarcina archaeal genus (49.19%). The data demonstrated that the performances of plants with identical design, in similar operating conditions, yielding comparable amount of biogas (average of 7237 m3 day−1 and 7916 m3 day−1 respectively for P1 and P2), VFA (1643 mg L−1 and 1634 mg L−1) and energy recovery (23.90–24 MWh day−1), depend on the stabilization of effective and functionally optimized methanogenic communities, but these communities are taxonomically different in the two biodigesters.
Graphical Abstract







Similar content being viewed by others
Data Availability
Data are available within the article. NGS and conventional sequences are available in GenBank (https://www.ncbi.nlm.nih.gov/genbank/).
References
Marañón, E., Salter, A.M., Castrillón, L., Heaven, S., Fernández-Nava, Y.: Reducing the environmental impact of methane emissions from dairy farms by anaerobic digestion of cattle waste. Waste Manag. 31, 1745–1751 (2011)
Stahel, W.R.: The circular economy. Nature 531, 435–438 (2016). https://doi.org/10.1038/531435a
Blades, L., Morgan, K., Douglas, R., Glover, S., De Rosa, M., Cromie, T., Smyth, B.: Circular Biogas-based economy in a rural agricultural setting. Energy Procedia 123, 89–96 (2017). https://doi.org/10.1016/j.egypro.2017.07.255
Benato, A., Macor, A.: Italian biogas plants: trend, subsidies, cost biogas composition and engine emissions. Energies 12(6), 979 (2019). https://doi.org/10.3390/en12060979
Rivière, D., Desvignes, V., Pelletier, E., Chaussonnerie, S., Guermazi, S., Weissenbach, J., Li, T., Camacho, P., Sghir, A.: Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 3, 700–714 (2009)
Tao, Y., Ersahin, M.E., Ghasimi, D.S.M., Ozgun, H., Wang, H., Zhang, X., Guo, M., Yang, Y., Stuckey, D.C., van Lier, J.B.: Biogas productivity of anaerobic digestion process is governed by a core bacterial microbiota. Chem. Eng. J. 380, 122425 (2020). https://doi.org/10.1016/j.cej.2019.122425
Mei, R., Nobu, M.K., Narihiro, T., Kuroda, K., Muñoz Sierra, J., Wu, Z., Ye, L., Lee, P.K.H., Lee, P.H., van Lier, J.B., McInerney, M.J., Kamagata, Y., Liu, W.T.: Operation-driven heterogeneity and overlooked feed-associated populations in global anaerobic digester microbiome. Water Res. 124, 77–84 (2017). https://doi.org/10.1016/j.watres.2017.07.050
Enzmann, F., Mayer, F., Rother, M., Holtmann, D.: Methanogens: biochemical background and biotechnological applications. AMB Expr. 8, 1 (2018). https://doi.org/10.1186/s13568-017-0531-x.
Conrad, R.: Importance of hydrogenotrophic, aceticlastic and methylotrophic methanogenesis for methane production in terrestrial, aquatic and other anoxic environments: a mini review. Pedosphere 30, 25–39 (2020). https://doi.org/10.1016/S1002-0160(18)60052-9
Yekta, S.S., Ziels, R.M., Björn, A., Skyllberg, U., Ejlertsson, J., Karlsson, A., Svedlund, M., Willén, M., Svensson, B.H.: Importance of sulfide interaction with iron as regulator of the microbial community in biogas reactors and its effect on methanogenesis, volatile fatty acids turnover, and syntrophic long-chain fatty acids degradation. J. Biosci. Bioeng. 123(5), 597–605 (2017). https://doi.org/10.1016/j.jbiosc.2016.12.003
Dai, X.H., Hu, C.L., Zhang, D., Dai, L.L., Duan, N.N.: Impact of a high ammonia- ammonium-pH system on methane-producing archaea and sulfate-reducing bacteria in mesophilic anaerobic digestion. Bioresour. Technol. 245, 598–605 (2017). https://doi.org/10.1016/j.biortech.2017.08.208
Yang, Z., Wang, W., He, Y., Zhang, R., Liu, G.: Effect of ammonia on methane production, methanogenesis pathway, microbial community and reactor performance under mesophilic and thermophilic conditions. Renew. Energy 125, 915–925 (2018). https://doi.org/10.1016/j.renene.2018.03.032
Raskin, L., Zheng, D., Griffin, M.E., Stroot, P.G., Misra, P.: Characterization of microbial communities in anaerobic bioreactors using molecular probes. Antonie Van Leeuwenhoek 68, 297–308 (1995). https://doi.org/10.1007/BF00874140
Krause, L., Diaz, N.N., Edwards, R.A., Gartemann, K.H., Krömeke, H., Neuweger, H., Pühler, A., Runte, K.J., Schlüter, A., Stoye, J., Szczepanowski, R., Tauch, A., Goesmann, A.: Taxonomic composition and gene content of a methane-producing microbial community isolated from a biogas reactor. J. Biotechnol. 136, 91–101 (2008). https://doi.org/10.1016/j.jbiotec.2008.06.003
Klocke, M., Nettmann, E., Bergmann, I., Mundt, K., Souidi, K., Mumme, J., Linke, B.: Characterization of the methanogenic Archaea within two-phase biogas reactor systems operated with plant biomass. Syst. Appl. Microbiol. 31, 190–205 (2008). https://doi.org/10.1016/j.syapm.2008.02.003
Yu, Y., Lee, C., Kim, J., Hwang, S.: Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 89, 670–679 (2005). https://doi.org/10.1002/bit.20347
Sundberg, C., Al-Soud, W.A., Larsson, M., Alm, E., Yekta, S.S., Svensson, B.H., Sørensen, S.J., Karlsson, A.: 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 85, 612–626 (2013). https://doi.org/10.1111/1574-6941.12148
Ma, S., Jiang, F., Huang, Y., Zhang, Y., Wang, S., Fan, H., Liu, B., Li, Q., Yin, L., Wang, H., Liu, H., Ren, Y., Li, S., Cheng, L., Fan, W., Deng. Y.: A microbial gene catalog of anaerobic digestion from full-scale biogas plants. Gigascience. 10, 1–10 (2021). https://doi.org/10.1093/gigascience/giaa164.
Luton, P.E., Wayne, J.M., Sharp, R.J., Riley, P.W.: The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148, 3521–3530 (2002). https://doi.org/10.1099/00221287-148-11-3521.14S
Rastogi, G., Ranade, D.R., Yeole, T.Y., Patole, M.S., Shouche, Y.S.: Investigation of methanogen population structure in biogas reactor by molecular characterization of methyl-coenzyme M reductase A (mcrA) genes. Bioresour. Technol. 99, 5317–5326 (2008). https://doi.org/10.1016/j.biortech.2007.11.024
Ranjard, L., Poly, F., Lata, J.C., Mougel, C., Thioulouse, J., Nazaret, S.: Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints. Appl. Environ. Microbiol. 67, 4479–4487 (2001). https://doi.org/10.1128/aem.67.10.4479-4487.2001
Hewson, I., Fuhrman, J.A.: Richness and diversity of bacterioplankton species along an estuarine gradient in Moreton Bay. Australia. Appl. Environ. Microbiol. 70, 3425–3433 (2004). https://doi.org/10.1128/AEM.70.6.3425-3433.2004
Cardinale, M., Brusetti, L., Quatrini, P., Borin, S., Puglia, A.M., Rizzi, A., Zanardini, E., Sorlini, C., Corselli, C., Daffonchio, D.: Comparison of different primer sets for use in automated ribosomal intergenic spacer analysis of complex bacterial communities. Appl. Environ. Microbiol. 70, 6147–6156 (2004). https://doi.org/10.1128/AEM.70.10.6147-6156.2004
Kovacs, A., Yacoby, K., Gophna, U.: A systematic assessment of automated ribosomal intergenic spacer analysis (ARISA) as a tool for estimating bacterial richness. Res. Microbiol. 161, 192–197 (2010). https://doi.org/10.1016/j.resmic.2010.01.006
Milani, C., Hevia, A., Foroni, E., Duranti, S., Turroni, F., Lugli, G.A., Sanchez, B., Martín, R., Gueimonde, M., van Sinderen, D., et al.: Assessing the fecal microbiota: an optimized ion torrent 16S rRNA gene-based analysis protocol. PLoS ONE 8, e68739 (2013). https://doi.org/10.1371/journal.pone.0068739
Fischer, M.A., Güllert, S., Neulinger, S.C., Streit, W.R., Schmitz, R.A.: Evaluation of 16S rRNA gene primer pairs for monitoring microbial community structures showed high reproducibility within and low comparability between datasets generated with multiple archaeal and bacterial primer pairs. Front. Microbiol. 7, 1297 (2016). https://doi.org/10.3389/fmicb.2016.01297
Caporaso, J.G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F.D., Costello, E.K., Fierer, N., Gonzalez Peña, A., Goodrich, J.K., Gordon, J.I., et al.: QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336 (2010). https://doi.org/10.1038/nmeth.f.303
Bokulich, N.A., Kaehler, B.D., Rideout, J.R., Dillon, M., Bolyen, E., Knight, R., Huttley, G.A., Caporaso, J.G.: Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6, 90 (2018). https://doi.org/10.1186/s40168-018-0470-z
Callahan, B.J., McMurdie, P.J., Rosen, M.J., Han, A.W., Johnson, A.J., Holmes, S.P.: DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583 (2016). https://doi.org/10.1038/nmeth.3869
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., Peplies, J., Glöckner, F.O.: The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013). https://doi.org/10.1093/nar/gks1219
Li, Y., Xu, H., Hua, D., Zhao, B., Mu, H., Jin, F., Meng, G., Fang, X.: Two-phase anaerobic digestion of lignocellulosic hydrolysate: focusing on the acidification with different inoculum to substrate ratios and inoculum sources. Sci. Total Environ. 699, 134226 (2020). https://doi.org/10.1016/j.scitotenv.2019.134226
Liu, T., Sun, L., Müller, B., Schnürer, A.: Importance of inoculum source and initial community structure for biogas production from agricultural substrates. Bioresour. Technol. 245, 768–777 (2017). https://doi.org/10.1016/j.biortech.2017.08.213
Cardinali-Rezende, J., Colturato, L.F.D.B., Colturato, T.D.B., Chartone-Souza, E., Nascimento, A.M.A., Sanz, J.L.: Prokaryotic diversity and dynamics in a full-scale municipal solid waste anaerobic reactor from start-up to steady-state conditions. Bioresour. Technol. 119, 373–383 (2012). https://doi.org/10.1016/j.biortech.2012.05.136
Kröber, M., Bekel, T., Diaz, N.N., Goesmann, A., Jaenicke, S., Krause, L., Miller, D., Runte, K.J., Viehöver, P., Pühler, A., Schlüter, A.: Phylogenetic characterization of a biogas plant microbial community integrating clone library 16S-rDNA sequences and metagenome sequence data obtained by 454-pyrosequencing. J. Biotechnol. 142, 38–49 (2009). https://doi.org/10.1016/j.jbiotec.2009.02.010
Lim, W., Ge, T., Tong, Y.W.: Monitoring of microbial communities in anaerobic digestion sludge for biogas optimisation. Waste Manag. 71, 334–341 (2018). https://doi.org/10.1016/j.wasman.2017.10.007
Calusinska, M., Goux, X., Fossépré, M., Muller, E.E.L., Wilmes, P., Delfosse, A.: A year of monitoring 20 mesophilic full-scale bioreactors reveals the existence of stable but different core microbiomes in bio-waste and wastewater anaerobic digestion systems. 11, 96 Biotechnol Biofuels. (2018). https://doi.org/10.1186/s13068-018-1195-8
Pelletier, E., Kreimeyer, A., Bocs, S., Rouy, Z., Gyapay, G., Chouari, R., Rivière, D., Ganesan, A., Daegelen, P., Sghir, A., Cohen, G.N., Médigue, C., Weissenbach, J., Le Paslier, D.: “Candidatus Cloacamonas acidaminovorans”: Genome sequence reconstruction provides a first glimpse of a new bacterial division. J. Bacteriol. 190, 2572–2579 (2008). https://doi.org/10.1128/JB.01248-07
Dyksma, S., Gallert, C.: Candidatus Syntrophosphaera thermopropionivorans: a novel player in syntrophic propionate oxidation during anaerobic digestion. Environ. Microbiol. Rep. 11, 558–570 (2019). https://doi.org/10.1111/1758-2229.12759
Conklin, A.S., Stensel, H.D., Ferguson, J.F.: The growth kinetics and competition between Methanosarcina and Methanosaeta in mesophilic anaerobic digestion. Proc. Water Environ. Fed. 2005, 100–121 (2012). https://doi.org/10.2175/193864705783867792
Karakashev, D., Batstone, D.J., Trably, E., Angelidaki, I.: Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 72, 5138–5141 (2006). https://doi.org/10.1128/AEM.00489-06
Powell, J.M., Broderick, G.A., Misselbrook, T.H.: Seasonal diet affects ammonia emissions from tie-stall dairy barns. J. Dairy Sci. 91, 857–869 (2008). https://doi.org/10.3168/jds.2007-0588
Minato, K., Kouda, Y., Yamakawa, M., Hara, S., Tamura, T., Osada, T.: Determination of GHG and ammonia emissions from stored dairy cattle slurry by using a floating dynamic chamber. Anim. Sci. J. 84, 165–177 (2013). https://doi.org/10.1111/j.1740-0929.2012.01053.x
Nobu, M.K., Narihiro, T., Kuroda, K., Mei, R., Liu, W.T.: Chasing the elusive Euryarchaeota class WSA2: genomes reveal a uniquely fastidious methyl-reducing methanogen. ISME J. 10, 2478–2487 (2016). https://doi.org/10.1038/ismej.2016.33
Wilkins, D., Lu, X.-Y., Shen, Z., Chen, J., Lee, P.K.H.: Pyrosequencing of mcrA and archaeal 16S rRNA genes reveals diversity and substrate preferences of methanogen communities in anaerobic digesters. Appl. Environ. Microbiol. 81, 604–613 (2015). https://doi.org/10.1128/aem.02566-14
Shakeri Yekta, S., Liu, T., Axelsson Bjerg, M., Ṥafarič, L., Karlsson, A., Björg, A., Schnürer, A.: Sulfide level in municipal sludge digesters affects microbial community response to long-chain fatty acid loads. Biotechnol. Biofuels. 12, 259–273 (2019). https://doi.org/10.1186/s13068-019-1598-1
Yang, J., Wang, D., Luo, Z., Zeng, W., Huang, H.: The role of reflux time in a leach bed reactor coupled with a methanogenic reactor for anaerobic digestion of pig manure: reactor performance and microbial community. J. Clean Prod. 242, 118367 (2020). https://doi.org/10.1016/j.jclepro.2019.118367
Ozbayram, E., Ince, O., Ince, B., Harms, H., Kleinsteuber, S.: Comparison of rumen and manure microbiomes and implications for the inoculation of anaerobic digesters. Microorganisms 6, 15 (2018). https://doi.org/10.3390/microorganisms6010015
Ciotola, R.J., Martin, J.F., Castańo, J.M., Lee, J., Michel, F.: Microbial community response to seasonal temperature variation in a small-scale anaerobic digester. Energies 6, 5182–5199 (2013). https://doi.org/10.3390/en6105182
Wahid, R., Mulat, D.G., Gaby, J.C., Horn, S.J.: Effects of H2:CO2 ratio and H2 supply fluctuation on methane content and microbial community composition during in-situ biological biogas upgrading. Biotechnol Biofuels 12, 104 (2019). https://doi.org/10.1186/s13068-019-1443-6
Lang, K., Schuldes, J., Klingl, A., Poehlein, A., Daniel, R., Brune, A.: New mode of energy metabolism in the seventh order of methanogens as revealed by comparative genome analysis of “Candidatus Methanoplasma termitum.” Appl. Environ. Microbiol. 81, 1338–1352 (2015). https://doi.org/10.1128/AEM.03389-14
Langer, S.G., Ahmed, S., Einfalt, D., Bengelsdorf, F.R., Kazda, M.: Functionally redundant but dissimilar microbial communities within biogas reactors treating maize silage in co-fermentation with sugar beet silage. Microb. Biotechnol. 8, 828–836 (2015)
Funding
This research was supported by funds to Prof. Anna Maria Sanangelantoni from FIL, of the University of Parma Local Funding for Research.
Author information
Authors and Affiliations
Contributions
CA: formal analysis, data curation, writing- original draft preparation. GV: validation, writing—review and editing. AMS: conceptualization, methodology, investigation, writing and reviewing. GF: sample and data collection.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Agrimonti, C., Visioli, G., Ferrari, G. et al. Comparison of Bacterial and Archaeal Microbiome in Two Bioreactors Fed with Cattle Sewage and Corn Biomass. Waste Biomass Valor 13, 4533–4547 (2022). https://doi.org/10.1007/s12649-022-01802-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12649-022-01802-0