Abstract
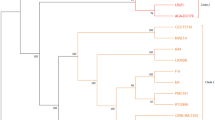
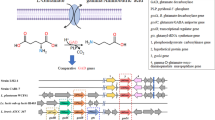
This study aimed to characterize the genomic and metabolic properties of a novel Lb. fermentum strain AGA52 which was isolated from a lactic acid fermented beverage called “shalgam.” The genome size of AGA52 was 2,001,184 bp, which is predicted to carry 2024 genes, including 50 tRNAs, 3 rRNAs, 3 ncRNAs, 15 CRISPR repeats, 14 CRISPR spacers, and 1 CRISPR array. The genome has a GC content of 51.82% including 95 predicted pseudogenes, 56 complete or partial transposases, and 2 intact prophages. The similarity of the clusters of orthologous groups (COG) was analyzed by comparison with the other Lb. fermentum strains. The detected resistome on the genome of AGA52 was found to be intrinsic originated. Besides, it has been determined that AGA52 has an obligate heterofermentative carbohydrate metabolism due to the absence of the 1-phosphofructokinase (pfK) enzyme. Furthermore, the strain is found to have a better antioxidant capacity and to be tolerant to gastrointestinal simulated conditions. It was also observed that the AGA52 has antimicrobial activity against Yersinia enterocolitica ATCC9610, Bacillus cereus ATCC33019, Salmonella enterica sv. Typhimurium, Escherichia coli O157:h7 ATCC43897, Listeria monocytogenes ATCC7644, Klebsiella pneumoniae ATCC13883, and Proteus vulgaris ATCC8427. Additionally, AGA52 exhibited 42.74 ± 4.82% adherence to HT29 cells. Cholesterol assimilation (33.9 ± 0.005%) and GABA production capacities were also confirmed by “in silico” and “in vitro.” Overall, the investigation of genomic and metabolic features of the AGA52 revealed that is a potential psychobiotic and probiotic dietary supplement candidate and can bring functional benefits to the host.








Similar content being viewed by others
References
Seddik HA, Bendali F, Gancel F, Fliss I, Spano G, Drider D (2017) Lactobacillus plantarum and its probiotic and food potentialities. Probiotics Antimicrob Proteins 9(2):111–122. https://doi.org/10.1007/s12602-017-9264-z
Buron-Moles G, Chailyan A, Dolejs I, Forster J, Miks MH (2019) Uncovering carbohydrate metabolism through a genotype-phenotype association study of 56 lactic acid bacteria genomes. Appl Microbiol Biotechnol 103(7):3135–3152. https://doi.org/10.1007/s00253-019-09701-6
Bangar SP, Suri S, Trif M, Ozogul F (2022) Organic acids production from lactic acid bacteria: a preservation approach. Food Biosci 46:101615
Tamang JP, Shin D-H, Jung S-J, Chae S-W (2016) Functional properties of microorganisms in fermented foods. Front Microbiol 7:578
Jiménez E, Langa S, Martín V, Arroyo R, Martín R, Fernández L, et al (2010) Complete genome sequence of Lactobacillus fermentum CECT 5716, a probiotic strain isolated from human milk. J Bacteriol Res 192(18):4800
Erol I, Kotil SE, Fidan O, Yetiman AE, Durdagi S, Ortakci F (2021) In silico analysis of bacteriocins from lactic acid bacteria against SARS-CoV-2. Probiotics Antimicrob 1–13
Sanders ME, Akkermans LM, Haller D, Hammerman C, Heimbach JT, Hörmannsperger G et al (2010) Safety assessment of probiotics for human use. Gut microbes 1(3):164–185
Zheng J, Wittouck S, Salvetti E, Franz CM, Harris H, Mattarelli P et al (2020) A taxonomic note on the genus Lactobacillus: description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int J Syst Evol Microbiol 70(4):2782–2858
D’ambrosio S, Ventrone M, Fusco A, Casillo A, Dabous A, Cammarota M et al (2022) Limosilactobacillus fermentum from buffalo milk is suitable for potential biotechnological process development and inhibits Helicobacter pylori in a gastric epithelial cell model. Biotechnology Reports 34:e00732
de Araújo Henriques Ferreira G, Magnani M, Cabral L, Brandão LR, Noronha MF, de Campos Cruz J, et al (2022) Potentially probiotic Limosilactobacillus fermentum fruit-derived strains alleviate cardiometabolic disorders and gut microbiota impairment in male rats fed a high-Fat diet. Probiotics Antimicrob 14(2):349–59
de Luna Freire MO, do Nascimento LCP, de Oliveira KÁR, de Oliveira AM, dos Santos Lima M, Napoleão TH, et al (2021) Limosilactobacillus fermentum strains with claimed probiotic properties exert anti-oxidant and anti-inflammatory properties and prevent cardiometabolic disorder in female rats fed a high-fat diet. Probiotics Antimicrob 1–13
Bhathena J, Martoni C, Kulamarva A, Tomaro-Duchesneau C, Malhotra M, Paul A et al (2013) Oral probiotic microcapsule formulation ameliorates non-alcoholic fatty liver disease in Bio F1B Golden Syrian hamsters. PLoS ONE 8(3):e58394
Yamamoto N, Shoji M, Hoshigami H, Watanabe K, Takatsuzu T, Yasuda S, et al (2019) Antioxidant capacity of soymilk yogurt and exopolysaccharides produced by lactic acid bacteria. Biosci Microbiota Food Health 18–017
Yetiman AE, Keskin A, Darendeli BN, Kotil SE, Ortakci F, Dogan M (2022) Characterization of genomic, physiological, and probiotic features Lactiplantibacillus plantarum DY46 strain isolated from traditional lactic acid fermented shalgam beverage. Food Biosci 46:101499
Paulino do Nascimento LC, Lacerda DC, Ferreira DJS, de Souza EL, de Brito Alves JL (2022) Limosilactobacillus fermentum, current evidence on the antioxidant properties and opportunities to be exploited as a probiotic microorganism. Probiotics Antimicrob 1–20
Feng H, Yuan Y, Yang Z, Xing X-h, Zhang C (2021) Genome-wide genotype-phenotype associations in microbes. J Biosci Bioeng 132(1):1–8
Coskun F (2017) A traditional Turkish fermented non-alcoholic beverage, “shalgam.” Beverages 3(4):49
Ekinci FY, Baser GM, Özcan E, Üstündağ ÖG, Korachi M, Sofu A et al (2016) Characterization of chemical, biological, and antiproliferative properties of fermented black carrot juice, shalgam. Eur Food Res Technol 242(8):1355–1368
Tanguler H, Erten H (2012) Occurrence and growth of lactic acid bacteria species during the fermentation of shalgam (salgam), a traditional Turkish fermented beverage. LWT-Food Science and Technology 46(1):36–41
Agirman B, Settanni L, Erten H (2021) Effect of different mineral salt mixtures and dough extraction procedure on the physical, chemical and microbiological composition of Şalgam: a black carrot fermented beverage. Food Chem 344:128618
Tanguler H, Erten H (2012) Chemical and microbiological characteristics of shalgam (şalgam): a traditional turkish lactic acid fermented beverage. J Food Qual 35(4):298–306
Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A (2020) Using SPAdes de novo assembler. Curr Protoc Bioinformatics 70(1):e102
Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L et al (2016) NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44(14):6614–6624
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C et al (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45(D1):D535–D542
Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ et al (2015) RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5(1):1–6
Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. https://doi.org/10.1186/1471-2164-12-402
Lee I, Kim YO, Park S-C, Chun J (2016) OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int J Syst Evol Microbiol 66(2):1100–1103
Chen I-MA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M et al (2021) The IMG/M data management and analysis system v. 6.0: new tools and advanced capabilities. Nucleic Acids Res 49(D1):D751-D63
Kanehisa M, Sato Y, Morishima K (2016) BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428(4):726–731
Zhang H, Yohe T, Huang L, Entwistle S, Wu P, Yang Z et al (2018) dbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res 46(W1):W95–W101
Wu S, Zhu Z, Fu L, Niu B, Li W (2011) WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics 12(1):1–9
Khan A, Mathelier A (2017) Intervene: a tool for intersection and visualization of multiple gene or genomic region sets. BMC Bioinformatics 18(1):1–8
Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y et al (2016) PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44(W1):W16–W21
Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V et al (2020) ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75(12):3491–500
Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard M, Edalatmand A et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1):D517–D525
Khan ZA, Siddiqui MF, Park S (2019) Current and emerging methods of antibiotic susceptibility testing. Diagnostics 9(2):49
CLSI C (2012) Performance standards for antimicrobial susceptibility testing; Twenty-Second Informational Supplement
Angmo K, Kumari A, Bhalla TC (2016) Probiotic characterization of lactic acid bacteria isolated from fermented foods and beverage of Ladakh. LWT-food Science and Technology 66:428–435
Zhang L, Ma H, Kulyar MF-e-A, Pan H, Li K, Li A et al (2022) Complete genome analysis of Lactobacillus fermentum YLF016 and its probiotic characteristics. Microb Pathog 62:105212
Krausova G, Hyrslova I, Hynstova I (2019) In vitro evaluation of adhesion capacity, hydrophobicity, and auto-aggregation of newly isolated potential probiotic strains. Fermentation 5(4):100
Mishra V, Prasad D (2005) Application of in vitro methods for selection of Lactobacillus casei strains as potential probiotics. Int J Food Microbiol 103(1):109–115
Ozturk G, Yetiman AE, Dogan M (2019) The bioactive efficiency of ultrasonic extracts from acorn leaves and green walnut husks against Bacillus cereus: a hybrid approach to PCA with the Taguchi method. Journal of Food Measurement and Characterization 13(2):1257–1268
Pieniz S, Andreazza R, Anghinoni T, Camargo F, Brandelli A (2014) Probiotic potential, antimicrobial and antioxidant activities of Enterococcus durans strain LAB18s. Food Control 37:251–256
Rudel LL, Morris M (1973) Determination of cholesterol using o-phthalaldehyde. J Lipid Res 14(3):364–366
Brandt K, Nethery MA, O’Flaherty S, Barrangou R (2020) Genomic characterization of Lactobacillus fermentum DSM 20052. BMC Genomics 21(1):1–13
Juhas M, Van Der Meer JR, Gaillard M, Harding RM, Hood DW, Crook DW (2009) Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol Rev 33(2):376–393
Yoo D, Bagon BB, Valeriano VDV, Oh JK, Kim H, Cho S et al (2017) Complete genome analysis of Lactobacillus fermentum SK152 from kimchi reveals genes associated with its antimicrobial activity. FEMS Microbiol Lett 364(18)
El-Ghaish S, Dalgalarrondo M, Choiset Y, Sitohy M, Ivanova I, Haertlé T et al (2010) Characterization of a new isolate of Lactobacillus fermentum IFO 3956 from Egyptian Ras cheese with proteolytic activity. Eur Food Res Technol 230(4):635–643
Duar RM, Lin XB, Zheng J, Martino ME, Grenier T, Pérez-Muñoz ME et al (2017) Lifestyles in transition: evolution and natural history of the genus Lactobacillus. FEMS Microbiol Rev 41(Supp_1):S27-S48
Cox AJ, Pyne DB, Saunders PU, Fricker PA (2010) Oral administration of the probiotic Lactobacillus fermentum VRI-003 and mucosal immunity in endurance athletes. Br J Sports Med 44(4):222–226
Hammes WP, Hertel C (2006) The genera Lactobacillus and Carnobacterium. Prokaryotes. Springer;p. 320–403
Vellend M (2010) Conceptual synthesis in community ecology. Q Rev Biol 85(2):183–206
Goh YJ, Barrangou R (2019) Harnessing CRISPR-Cas systems for precision engineering of designer probiotic lactobacilli. Curr Opin Biotechnol 56:163–171
Roberts A, Barrangou R (2020) Applications of CRISPR-Cas systems in lactic acid bacteria. FEMS Microbiol Rev 44(5):523–537
Hidalgo-Cantabrana C, O’Flaherty S, Barrangou R (2017) CRISPR-based engineering of next-generation lactic acid bacteria. Curr Opin Microbiol 37:79–87
Hacker J, Kaper JB (2000) Pathogenicity islands and the evolution of microbes. Annual Reviews in Microbiology 54(1):641–679
Oechslin F, Zhu X, Dion MB, Shi R, Moineau S (2022) Phage endolysins are adapted to specific hosts and are evolutionarily dynamic. PLoS Biol 20(8):e3001740
Miller-Ensminger T, Mormando R, Maskeri L, Shapiro JW, Wolfe AJ, Putonti C (2020) Introducing Lu-1, a novel Lactobacillus jensenii phage abundant in the urogenital tract. PLoS ONE 15(6):e0234159
Cahill J, Young R (2019) Phage lysis: multiple genes for multiple barriers. Adv Virus Res 103:33–70
Kleerebezem M, Boekhorst J, van Kranenburg R, Molenaar D, Kuipers OP, Leer R et al (2003) Complete genome sequence of Lactobacillus plantarum WCFS1. Proc Natl Acad Sci 100(4):1990–1995
EFSA (2012) Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10(6):2740
Deghorain M, Goffin P, Fontaine L, Mainardi J-L, Daniel R, Errington J et al (2007) Selectivity for D-lactate incorporation into the peptidoglycan precursors of Lactobacillus plantarum: role of Aad, a VanX-like D-alanyl-D-alanine dipeptidase. J Bacteriol 189(11):4332–4337
Campedelli I, Mathur H, Salvetti E, Clarke S, Rea MC, Torriani S et al (2019) Genus-wide assessment of antibiotic resistance in Lactobacillus spp. Appl Environ Microbiol 85(1):e01738-e1818
Zhang Z-Y, Liu C, Zhu Y-Z, Wei Y-X, Tian F, Zhao G-P et al (2012) Safety assessment of Lactobacillus plantarum JDM1 based on the complete genome. Int J Food Microbiol 153(1–2):166–170
Rozman V, Lorbeg PM, Accetto T, Matijašić BB (2020) Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int J Food Microbiol 314:108388
Walter J, Heng NC, Hammes WP, Loach DM, Tannock GW, Hertel C (2003) Identification of Lactobacillus reuteri genes specifically induced in the mouse gastrointestinal tract. Appl Environ Microbiol 69(4):2044–2051
Gänzle MG, Follador R (2012) Metabolism of oligosaccharides and starch in lactobacilli: a review. Front Microbiol 3:340
Salvetti E, Harris HM, Felis GE, O’Toole PW (2018) Comparative genomics of the genus Lactobacillus reveals robust phylogroups that provide the basis for reclassification. Appl Environ Microbiol 84(17):e00993-e1018
Morita H, Toh H, Fukuda S, Horikawa H, Oshima K, Suzuki T et al (2008) Comparative genome analysis of Lactobacillus reuteri and Lactobacillus fermentum reveal a genomic island for reuterin and cobalamin production. DNA Res 15(3):151–161
Schleif R (2010) AraC protein, regulation of the l-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol Rev 34(5):779–796
Jiang T, Guo X, Yan J, Zhang Y, Wang Y, Zhang M et al (2017) A bacterial multidomain NAD-independent d-lactate dehydrogenase utilizes flavin adenine dinucleotide and Fe-S clusters as cofactors and quinone as an electron acceptor for d-lactate oxidization. J Bacteriol 199(22):e00342-e417
Thomas LM, Harper AR, Miner WA, Ajufo HO, Branscum KM, Kao L et al (2013) Structure of Escherichia coli AdhP (ethanol-inducible dehydrogenase) with bound NAD. Acta Crystallogr, Sect F: Struct Biol Cryst Commun 69(7):730–732
Goel A, Halami PM, Tamang JP (2020) Genome analysis of Lactobacillus plantarum isolated from some Indian fermented foods for bacteriocin production and probiotic marker genes. Front Microbiol 40
Martienssen M, O R, U K (2001) Surface properties of bacteria from different wastewater treatment plants. Acta Biotechnol 21(3):207–25
de Souza BMS, Borgonovi TF, Casarotti SN, Todorov SD, Penna ALB (2019) Lactobacillus casei and Lactobacillus fermentum strains isolated from mozzarella cheese: probiotic potential, safety, acidifying kinetic parameters and viability under gastrointestinal tract conditions. Probiotics and antimicrobial proteins 11(2):382–396
Chaffanel F, Charron-Bourgoin F, Soligot C, Kebouchi M, Bertin S, Payot S et al (2018) Surface proteins involved in the adhesion of Streptococcus salivarius to human intestinal epithelial cells. Appl Microbiol Biotechnol 102(6):2851–2865
Pan M, Kumaree KK, Shah NP (2017) Physiological changes of surface membrane in Lactobacillus with prebiotics. J Food Sci 82(3):744–750
Haddaji N, Mahdhi AK, Krifi B, Ismail MB, Bakhrouf A (2015) Change in cell surface properties of Lactobacillus casei under heat shock treatment. FEMS Microbiol Lett 362(9)
Lesuffleur T, Barbat A, Dussaulx E, Zweibaum A (1990) Growth adaptation to methotrexate of HT-29 human colon carcinoma cells is associated with their ability to differentiate into columnar absorptive and mucus-secreting cells. Can Res 50(19):6334–6343
Li Q, Liu X, Dong M, Zhou J, Wang Y (2015) Aggregation and adhesion abilities of 18 lactic acid bacteria strains isolated from traditional fermented food. Int J Agric Policy Res 3(2):84–92
Ouwehand AC, Kirjavainen PV, Shortt C, Salminen S (1999) Probiotics: mechanisms and established effects. Int Dairy J 9(1):43–52
Saarela M, Mogensen G, Fonden R, Mättö J, Mattila-Sandholm T (2000) Probiotic bacteria: safety, functional and technological properties. J Biotechnol 84(3):197–215
Gao Y, Liu Y, Sun M, Zhang H, Mu G, Tuo Y (2020) Physiological function analysis of Lactobacillus plantarum Y44 based on genotypic and phenotypic characteristics. J Dairy Sci 103(7):5916–5930
Fei Y, Li L, Zheng Y, Liu D, Zhou Q, Fu L (2018) Characterization of Lactobacillus amylolyticus L6 as potential probiotics based on genome sequence and corresponding phenotypes. LWT 90:460–468
Tomaro-Duchesneau C, Jones ML, Shah D, Jain P, Saha S, Prakash S (2014) Cholesterol assimilation by Lactobacillus probiotic bacteria: an in vitro investigation. Biomed Res Int 2014
Pan DD, Zeng XQ, Yan YT (2011) Characterisation of Lactobacillus fermentum SM-7 isolated from koumiss, a potential probiotic bacterium with cholesterol-lowering effects. J Sci Food Agric 91(3):512–518
Cui Y, Miao K, Niyaphorn S, Qu X (2020) Production of gamma-aminobutyric acid from lactic acid bacteria: a systematic review. Int J Mol Sci 21(3):995
Wu J-Y, Matsuda T, Roberts E (1973) Purification and characterization of glutamate decarboxylase from mouse brain. J Biol Chem 248(9):3029–3034
Okada T, Sugishita T, Murakami T, Murai H, Saikusa T, Horino T, et al (2000) Effect of the defatted rice germ enriched with GABA for sleeplessness, depression, autonomic disorder by oral administration. Nippon Shokuhin Kagaku Kogaku Kaishi 47(8):596–603
Abdou AM, Higashiguchi S, Horie K, Kim M, Hatta H, Yokogoshi H (2006) Relaxation and immunity enhancement effects of γ-aminobutyric acid (GABA) administration in humans. BioFactors 26(3):201–208
Ting Wong CG, Bottiglieri T, Snead OC III (2003) Gaba, γ-hydroxybutyric acid, and neurological disease. Ann Neurol: Official Journal of the American Neurological Association and the Child Neurology Society 54(S6):S3–S12
Hayakawa K, Kimura M, Kasaha K, Matsumoto K, Sansawa H, Yamori Y (2004) Effect of a γ-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br J Nutr 92(3):411–417
Leventhal AG, Wang Y, Pu M, Zhou Y, Ma Y (2003) GABA and its agonists improved visual cortical function in senescent monkeys. Science 300(5620):812–815
Lyu C, Zhao W, Peng C, Hu S, Fang H, Hua Y et al (2018) Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb Cell Fact 17(1):1–14
Wu Q, Tun HM, Law Y-S, Khafipour E, Shah NP (2017) Common distribution of gad operon in Lactobacillus brevis and its GadA contributes to efficient GABA synthesis toward cytosolic near-neutral pH. Front Microbiol 8:206
Yunes R, Poluektova E, Dyachkova M, Klimina K, Kovtun A, Averina O et al (2016) GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe 42:197–204
Acknowledgements
Mr. Mehmet Horzum would like to thank the Council of Higher Education of Türkiye (YÖK) due to the 100/2000 PhD scholarship program.
Funding
This study has been financially supported by Erciyes University Scientific Research Projects Coordination Unit under grant number FKB-2020–10551.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study’s conception and design. Material preparation, data collection, and analysis were performed by Ahmet E. Yetiman, Mehmet Horzum, and Dilek Bahar. The first draft of the manuscript was written by Ahmet E. Yetiman and was proofread by Mikail Akbulut. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yetiman, A., Horzum, M., Bahar, D. et al. Assessment of Genomic and Metabolic Characteristics of Cholesterol-Reducing and GABA Producer Limosilactobacillus fermentum AGA52 Isolated from Lactic Acid Fermented Shalgam Based on “In Silico” and “In Vitro” Approaches. Probiotics & Antimicro. Prot. 16, 334–351 (2024). https://doi.org/10.1007/s12602-022-10038-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-022-10038-2