Abstract
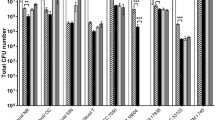
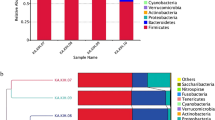
In this study, we describe enhanced in vitro probiotic activities of preformed biofilms versus planktonic cultures of Lactobacillus fermentum LfQi6 (LfQi6), a lactic acid bacterium (LAB) isolated from the human microbiome. These evaluations are used to help predict host in vivo probiotic benefits and therefore indicate that LfQi6 may provide significant probiotic benefits in the human host when administered as preformed biofilms rather than as planktonic cultures. Specifically, LfQi6 biofilms demonstrated improved in vitro performance versus LfQi6 planktonic cultures for host gastrointestinal survival and engraftment, strain-specific antimicrobial and anti-biofilm activity against clinically significant pathogens, concurrent promotion of beneficial gastrointestinal commensal biofilms, beneficial commensal enzyme activities, and host cellular-protective glutathione antioxidant activity. Evaluation of LfQi6 according to the European Food Safety Authority (EFSA 2007, 2012, 2015) Guidelines and Joint FAO/WHO Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food Evaluation of Probiotics in Food (FAO/WHO, 2002) demonstrates strain safety. In summary, in vitro evaluation of Lact. fermentum LfQi6 demonstrates significant evidence for strain-specific probiotic characteristics and safety. Moreover, strain-specific as well as biofilm-phenotype-specific benefits demonstrated in vitro furthermore suggest that in vivo use of LfQi6 biofilm biomass may be of greater benefit to the human host than the use of standard planktonic cultures. This concept – potentiating probiotic benefits through the use of preformed commensal biofilms – is novel and may serve to further broaden the application of microbial biofilms to human health.









Similar content being viewed by others
References
FAO/WHO (2001) Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Report of the Joint Food and Agriculture (FAO) of the United Nations/World Health Organization (WHO) Expert Consultation on Evaluation of Health and Nutritional Properties of Probiotics in Food including Powder Milk with Live Lactic Acid Bacteria
Szajewska H, Setty M, Mrukowicz J, Guandalini S (2006) Probiotics in gastrointestinal diseases in children: hard and not-so-hard evidence of efficacy. J Pediatr Gastroenterol Nutr 42(5):454–475. https://doi.org/10.1097/01.mpg.0000221913.88511.72
Szajewska H, Ruszczynski M, Radzikowski A (2006) Probiotics in the prevention of antibiotic-associated diarrhea in children: a meta-analysis of randomized controlled trials. J Pediatr 149(3):367–372. https://doi.org/10.1016/j.jpeds.2006.04.053
Costerton JW, Stewart PS, Greenberg EP (1999) Bacterial biofilms: a common cause of persistent infections. Science 284(5418):1318–1322. https://doi.org/10.1126/science.284.5418.1318
Costerton JW, Veeh R, Shirtliff M, Pasmore M, Post C, Ehrlich G (2003) The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 112(10):1466–1477. https://doi.org/10.1172/jci20365
Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15(12):167–193. https://doi.org/10.1128/cmr.15.2.167-193.2002
Subhadra B, Krier J, Hofstee K, Monsul N, Berkes E (2015) Draft whole-genome sequence of Lactobacillus fermentum LfQi6, derived from the human microbiome. Genome Announc 3(3):e00423–e00415. https://doi.org/10.1128/genomeA.00423-15
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam HF, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R (2010) A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 38(Web Server issue):W695–W699. https://doi.org/10.1093/nar/gkq313
Dewey CN (2012) Whole-genome alignment. Methods Mol Biol 855:237–257. https://doi.org/10.1007/978-1-4939-9074-0_4
Galia E, Nicoliades E, Horter D, Lobenberg R, Reppas C, Dressman JB (1998) Evaluation of various dissolution media for predicting in vivo performance of class I and II drugs. Pharm Res 15(5):698–705
Rosenberg M (1984) Bacterial adherence to hydrocarbons: a useful technique for studying cell surface hydrophobicity. FEMS Microbiol Lett 22(3):289–295. https://doi.org/10.1111/j.1574-6968.1984.tb00743.x
Wiegand I, Hilpert K, Hancock RE (2008) Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat Protoc 3(2):163–175. https://doi.org/10.1038/nprot.2007.521
EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP) (2012) Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA J 10:2740 http://onlinelibrary.wiley.com/doi/10.2903/sp.efsa.2018.EN-1389/full. Accessed 24 Jul 18
Bover-Cid S, Holzapfel WH (1999) Improved screening procedure for biogenic amine production by lactic acid bacteria. Int J Food Microbiol 53(1):33–41. https://doi.org/10.1016/S0168-1605(99)00152-X
Zhou JS, Gopal PK, Gill HS (2001) Potential probiotic lactic acid bacteria Lactobacillus rhamnosus (HN001), Lactobacillus acidophilus (HN017) and Bifidobacterium lactis (HN019) do not degrade gastric mucin in vitro. Int J Food Microbiol 63(1–2):81–90. https://doi.org/10.1016/S0168-1605(00)00398-6
Wattam AR, Davis JJ, Assaf R, Boisvert S, Brettin T, Bun C, Conrad N, Dietrich EM, Disz T, Gabbard JL, Gerdes S, Henry CS, Kenyon RW, Machi D, Mao C, Nordberg EK, Olsen GJ, Murphy-Olson DE, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Vonstein V, Warren A, Xia F, Yoo H, Stevens RL (2017) Improvements to PATRIC, the all-bacterial bioinformatics database and analysis resource center. Nucleic Acids Res 45(D1):D535–D542. https://doi.org/10.1093/nar/gkw1017
Reuter G (1965) Das vorkommen on laktobazillen in lebensmittel und ihr verhalten im menschlichen intestinaltrakt. Zentbl Bakteriol Parasitol Infekt Hyg I Orig 197S:468–487
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Sahadeva RPK, Leong SF, Chua KH, Tan CH, Chan HY, Tong EV, Wong SYW, Chan HK (2011) Survival of commercial probiotic strains to pH and bile. Int Food Res J 18(4):1515–1522 http://www.ifrj.upm.edu.my/18%20(04)%202011/(44)IFRJ-2011-285.pdf. Accessed 26 Feb 19
Chou LS, Weimer B (1999) Isolation and characterization of acid- and bile-tolerant isolates from strains of Lactobacillus acidophilus. J Dairy Sci 82(1):23–31. https://doi.org/10.3168/jds.S0022-0302(99)75204-5
Overbeek R, Olson R, Pusch GD, Olson GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R (2014) The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42(Database Issue):D206–D214. https://doi.org/10.1093/nar/gkt1226
Pophaly SD, Singh R, Pophaly SD, Kaushik JK, Tomar SK (2012) Current status and emerging role of glutathione in food grade lactic acid bacteria. Microb Cell Factories 11:114. https://doi.org/10.1186/1475-2859-11-114
Nikolic M, Jovcic B, Kojic M, Topisirovic L (2010) Surface properties of Lactobacillus and Leuconostoc isolates from homemade cheeses showing auto-aggregation ability. Eur Food Res Technol 231(6):925–931. https://doi.org/10.1007/s00217-101-1344-1
Hussain M, Khan NT, Wajid A, Rasool SA (2008) Technological characterization of indigenous enterococcal population for probiotic potential. Pak J Bot 40:2
Thongaram T, Hoeflinger J, Chow J, Miller MJ (2017) Prebiotic galactooligosaccharide metabolism by probiotic lactobacilli and bifidobacteria. J Agric Food Chem 65(20):4184–4192. https://doi.org/10.3168/jds.2017-12753
LeBlanc JG, Ledue-Clier F, Bensaada M, de Giori GS, Guerekobaya T, Sesama F, Juillard V, Rabot S, Piard JC (2008) Ability of Lactobacillus fermentum to overcome host alpha-galactosidase deficiency, as evidenced by reduction of hydrogen excretion in rats consuming soya alpha-galacto-oligosaccharides. BMC Microbiol 8:22. https://doi.org/10.1186/1471-2180-8-22
European Food Safety Authority (EFSA) (2008) Update of the criteria used in the assessment of bacterial resistance to antibiotics of human or veterinary importance. EFSA J 732:1–15
Costerton JW, Cheng KJ, Geesey GG, Ladd TI, Nickel JC, Dasgupta M, Marrie TJ (1987) Bacterial biofilms in nature and disease. Ann Rev Microbiol 41:435–464. https://doi.org/10.1146/annurev.mi.41.100187.002251
Palestrant D, Holzknecht ZE, Collins BH, Parker W, Miller SE, Bollinger RR (2004) Microbial biofilms in the gut: visualization by electron microscopy and by acridine orange staining. Ultrastruct Pathol 28(1):23–27. https://doi.org/10.1080/01913120490275196
Linnes JC, Ma H, Bryers JD (2013) Giant extracellular matrix binding protein expression in Staphylococcus epidermidis is regulated by biofilm formation and osmotic pressure. Curr Microbiol 66(6):627–633. https://doi.org/10.1007/s00284-013-0316-7
Rubio R, Jofre A, Martin B, Aymerich T, Garriga M (2014) Characterization of lactic acid bacteria isolated from infant faeces as potential probiotic starter cultures for fermented sausages. Food Microbiol 38:303–311. https://doi.org/10.1016/j.fm.2013.07.015
Ferrnandez MF, Boris S, Barbes C (2003) Probiotic properties of human lactobacilli strains to be used in the gastrointestinal tract. J Appl Microbiol 94(3):449–455. https://doi.org/10.1046/j.1365-2672.2003.01850.x
Prasad J, Gill HS, Smart J, Gopal PK (1998) Selection and characterization of Lactobacillus and Bifidobacterium strains for use as probiotics. Int Dairy J 8(12):993–1002. https://doi.org/10.1016/S0958-6946(99)00024-2
Islam MA, Bajracharya P, Kang SK, Yun CH, Kim EM, Jeong HJ, Choi YJ, Kim EB, Cho CS (2011) Mucoadhesive alginate/poly (L-lysine)/thiolated alginate microcapsules for oral delivery of Lactobacillus salivarius 29. J Nanosci Nanotechnol 11(8):7091–7095. https://doi.org/10.1166/jnn.2011.4858
Ruiz L, Margolles A, Sanchez B (2013) Bile resistance mechanisms in Lactobacillus and Bifidobacterium. Front Microbiol 4:396. https://doi.org/10.3389/fmicb.2013.00396
Jones BV, Begley M, Hill C, Gahan CG, Marchesi JR (2008) Functional and comparative metagenomic analysis of bile salt hydrolase activity in the human gut microbiome. Proc Natl Acad Sci U S A 105(36):13580–13585. https://doi.org/10.1073/pnas.0804437105
Begley M, Hill C, Gahan CG (2006) Bile salt hydrolase activity in probiotics. Appl Environ Microbiol 72(3):1729–1738. https://doi.org/10.1128/aem.72(3):1729-1738.2006
Miremadi F, Ayyash M, Sherkat F, Stojanovska L (2014) Cholesterol reduction mechanisms and fatty acid composition of cellular membranes of probiotic Lactobacilli and Bifidobacteria. J Funct Foods 9:295–305. https://doi.org/10.1016/j.jff.2014.05.002
Dong Z, Zhang J, Lee B, Li H, Du G, Chen J (2012) A bile salt hydrolase gene of Lactobacillus plantarum BBE7 with cholesterol-removing activity. Eur Food Res Technol 235(3):419–427. https://doi.org/10.1007/s00217-012-1769-9
Noriega L, Cuevas I, Margolles A, de los Reyes-Gavilan CG (2006) Deconjugation and bile salts hydrolase activity by Bifidobacterium strains with acquired resistance to bile. Int Dairy J 16(8):850–855. https://doi.org/10.1016/j.idairyj.2005.09.008
Klaver FA, van der Meer R (1993) The assumed assimilation of cholesterol by Lactobacilli and Bifidobacterium bifidum is due to their bile salt-deconjugating activity. Appl Environ Microbiol 59(4):1120–1124
Aoudia N, Rieu A, Briandet R, Deschamps J, Chluba J, Jego G, Garrido C, Guzzo J (2016) Biofilms of Lactobacillus plantarum and Lactobacillus fermentum: effect on stress responses, antagonistic effects on pathogen growth and immunomodulatory properties. Food Microbiol 53(Pt A):51–59. https://doi.org/10.1016/j.fm.2015.04.009
Bron PA, Molenaar D, de Vos WM, Kleerebezem M (2006) DNA micro-array-based of bile-responsive genes in Lactobacillus plantarum. J Appl Microbiol 100(4):728–738. https://doi.org/10.1111/j.1365-2672.2006.02891.x
Mikelsaar M, Zilmer M (2009) Lactobacillus fermentum ME-3 - an antimicrobial and antioxidative probiotic. Microb Ecol Health Dis 21(1):1–27. https://doi.org/10.1080/08910600902815561
Amaretti A, di Nunzio M, Pompei A, Raimondi S, Rossi M, Bordoni A (2013) Antioxidant properties of potentially probiotic bacteria: in vitro and in vivo activities. Appl Microbiol Biotechnol 97(2):809–817. https://doi.org/10.1007/s00253-012-4241-7
Mishra V, Shah C, Mokashe N, Chavan R, Yadav H, Prajapati J (2015) Probiotics as potential antioxidants: a systematic review. J Agric Food Chem 63(14):3615–3626. https://doi.org/10.1021/jf506326t
Wu G, Fang YZ, Yang S, Lupton JR, Turner ND (2004) Glutathione metabolism and its implications for health. J Nutr 134(3):489–492. https://doi.org/10.1093/jn/134.3.489
Jones DP (2002) Redox potential of GSH/GSSG couple: assay and biological significance. Methods Enzymol 348:93–112. https://doi.org/10.1016/s0076-6879(02)48630-2
Valencia E, Marina A, Hardy G (2001) Glutathione-nutritional and pharmacologic viewpoints: part IV. Nutrition 17(9):783–784
Copley SD, Dhillon JK (2002) Lateral gene transfer and parallel evolution in the history of glutathione biosynthesis genes. Genome Biol 3(5):research0025. https://doi.org/10.1186/gb-2002-3-5-research0025
Kullisaar T, Songisepp E, Aunapuu M, Kilk K, Arend A, Mikelsaar M, Rehema A, Zilmer M (2010) Complete glutathione system in probiotic Lactobacillus fermentum ME-3. Prikl Biokhim Mikrobiol 46(5):527–531
Stewart PS, Zhang T, Xu R, Pitts B, Walters MC, Roe F, Kikhney J, Moter A (2016) Reaction-diffusion theory explains hypoxia and heterogeneous growth within microbial biofilms associated with chronic infections. NPJ Biofilms Microbiomes 2:16012. https://doi.org/10.1038/npjbiofilms.2016.12
Stewart PS, Costerton JW (2001) Antibiotic resistance of bacteria in biofilms. Lancet 358(9276):135–138. https://doi.org/10.1016/s0140-6736(01)05321-1
Saarela M, Mogensen G, Fonden R, Matto J, Mattila-Sandholm T (2000) Probiotic bacteria: safety, functional and technological properties. J Biotechnol 84(3):197–215. https://doi.org/10.1016/S0168-1656(00)00375-8
Argyri AA, Zoumpopoulou G, Karatzas KA, Tsakalidou E, Nychas GJ, Panagou EZ, Tassou CC (2013) Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol 33(2):282–291. https://doi.org/10.1016/j.fm.2012.10.005
Krasowska A, Sigler K (2014) How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol 4:112. https://doi.org/10.3389/fcimb.2014.00112
Collado MC, Isolauri E, Salminen S (2008) Specific probiotic strains and their combinations counteract adhesion of Enterobacter sakazakii to intestinal mucus. FEMS Microbiol Lett 285(1):58–64. https://doi.org/10.1111/j.1574-6968.2008.01211.x
Keller MK, Hasslof P, Stecksen-Blicks C, Twetman S (2011) Co-aggregation and growth inhibition of probiotic lactobacilli and clinical isolates of mutans streptococci: an in vitro study. Acta Odontol Scand 69(5):263–268. https://doi.org/10.3109/00016357.2011.554863
Velez M, De Keersmaecker SC, Vanderleyden J (2007) Adherence factors of Lactobacillus in the human intestinal tract. FEMS Microbiol Lett 276(2):140–148. https://doi.org/10.1111/j.1574-6968.2007.00908.x
Fazary AE, Ju YH (2007) Feruloyl esterases as biotechnological tools: current and future perspectives. Acta Biochim Biophys Sin Shanghai 39(11):811–828. https://doi.org/10.1111/j.1745-7270.2007.00348.x
Bhathena J, Martoni C, Kulamarva A, Urbanska AM, Malhotra M, Prakash S (2009) Orally delivered microencapsulated live probiotic formulation lowers serum lipids in hypercholesterolemic hamsters. J Med Food 12(2):310–319. https://doi.org/10.1371/journal.pone.0058394
Srinivasan M, Sudheer AR, Menon VP (2007) Ferulic acid: therapeutic potential through its antioxidant property. J Clin Biochem Nutr 40(2):92–100. https://doi.org/10.3164/jcbn.40.92
Andreasen MF, Kroon PA, Williamson G, Garcia-Conesa MT (2001) Esterase activity able to hydrolyze dietary antioxidant hydroxycinnamates is distributed along the intestine of mammals. J Agric Food Chem 49(11):5679–5684. https://doi.org/10.1021/jf010668c
Mukdsi MC, Cano MP, Gonzalez SN, Medina RB (2012) Administration of Lactobacillus fermentum CRL1446 increases intestinal feruloyl esterase activity in mice. Lett Appl Microbiol 54(1):18–25. https://doi.org/10.1111/j.1472-765X.2011.03166.x
Bhathena J, Tomaro-Duchesneau C, Martoni C, Malhotra M, Kulamarva A, Urbanska MA, Paul A, Prakash S (2012) Effect of orally administered microencapsulated FA-producing Lact. fermentum on markers of metabolic syndrome: an in vivo analysis. J Diabetes Metab S6:006. https://doi.org/10.1089/jmf.2008.0166
Tomaro-Duchesneau C, Saha S, Malhotra M, Jones ML, Labbe A, Rodes L, Kahouli I, Prakash S (2014) Effect of orally administered Lact. fermentum NCIMB 5221 on markers of metabolic syndrome: an in vivo analysis using ZDF rats. Appl Microbiol Biotechnol 98(1):115–126. https://doi.org/10.1007/s00253-013-5252-8
Couteau D, McCartney AL, Gibson GR, Williamson G, Faulds CB (2001) Isolation and characterization of human colonic bacteria able to hydrolyse chlorogenic acid. J Appl Microbiol 90(6):873–881. https://doi.org/10.1046/j.1365-2672.2001.01316.x
Wasfi R, Abd El-Rahman OA, Zafer MM, Ashour HM (2018) Probiotic Lactobacillus sp. inhibit growth, biofilm formation and gene expression of caries-inducing Streptococcus mutans. J Cell Mol Med 22(3):1972–1983. https://doi.org/10.1111/jcmm
Chikindas ML, Weeks R, Drider D, Chistyakov VA, Dicks LM (2018) Functions and emerging applications of bacteriocins. Curr Opin Biotechnol 49:23–28. https://doi.org/10.1016/j.copbio.2017.07.011
Funding
This project is exclusively funded by Quorum Innovations LLC (Sarasota, Florida), a human microbiome-based therapeutics discovery biotechnology company.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Drs. Berkes and Monsul report the following disclosures: Co-founders of Quorum Innovations, LLC (Sarasota, FL). They are also co-inventors on several patents in the area of the human microbiome and biofilm modulation. The remaining authors are or were affiliated with Quorum Innovations, LLC.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Berkes, E., Liao, YH., Neef, D. et al. Potentiated In Vitro Probiotic Activities of Lactobacillus fermentum LfQi6 Biofilm Biomass Versus Planktonic Culture. Probiotics & Antimicro. Prot. 12, 1097–1114 (2020). https://doi.org/10.1007/s12602-019-09624-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-019-09624-8