Abstract
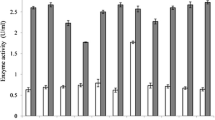
Probiotics are beneficial microorganisms and have long been used in food production as well as health promotion products. Bioengineered probiotics are used to express and transfer native or recombinant molecules to the mucosal surface of the digestive tract to improve feed efficiency and promote health. Lactococcus lactis is a potential probiotic candidate to produce useful biological proteins. The aim of this investigation was to develop a recombinant Lactococcus lactis with the potential of producing phytase. To enhance the efficiency of expression and secretion of recombinant phytase, usp45 signal peptide was added to the expression vector containing phytase gene (appA2) derived from Escherichia coli. Sequencing of recombinant plasmid containing appA2 showed the correct construction of plasmid. Total length of the phytase insert was 1.25 kbp. A Blast search of the cloned fragment showed 99% similarity to the reported E. coli phytase sequence in the GenBank (accession number: AM946981.2). A plasmid containing usp45 and appA2 electrotransferred into Lactococcus lactis. Zymogram with polyacrylamide gel revealed that the protein extract from the supernatant and the cell pellet of recombinant bacteria had phytase activity. Enzyme activity of 4 U/ml was obtained in cell extracts, and supernatant maximal phytase activity was 19 U/ml. The recombinant L. lactis was supplemented in broiler chicken feed and showed the increase of apparent digestibility on phytate phosphorus in the digestive tract and it was same as performance of E. coli commercial phytase.



Similar content being viewed by others
References
Graf E (1983) Applications of phytic acid. J Am Oil Chem Soc 60:1861–1867. https://doi.org/10.1007/BF02901539
Holm PB, Kristiansen KN, Pedersen HB (2002) Transgenic approaches in commonly consumed cereals to improve iron and zinc content and bioavailability. J Nutr 132(3):514S–516S. https://doi.org/10.1093/jn/132.3.514S
Wodzinski RJ, Ullah AH (1996) Phytase. Adv Appl Microbiol 42:263–302. https://doi.org/10.1016/S0065-2164(08)70375-7
Lei XG, Stahl CH (2001) Biotechnological development of effective phytases for mineral nutrition and environmental protection. Appl Microbiol Biotechnol 57(4):474–481. https://doi.org/10.1007/s002530100795
Mullaney EJ, Ullah AH (2003) The term phytase comprises several different classes of enzymes. Biochem Biophys Res Commun 312(1):179–184. https://doi.org/10.1016/j.bbrc.2003.09.176
Tufarelli V, Crovace AM, Rossi G, Laudadio V (2017) Effect of a dietary probiotic blend on performance, blood characteristics, meat quality and faecal microbial shedding in growing-finishing pigs. S Afr J Anim Sci 47(6):875–882. https://doi.org/10.4314/sajas.v47i6.15
Salminen S, Bouley C, Boutron-Ruault MC, Cummings JH, Franck A, Gibson GR, Isolauri E, Moreau MC, Roberfroid M, Rowland I (1998) Functional food science and gastrointestinal physiology and function. Br J Nutr 80(Suppl 1):S147–S171. https://doi.org/10.1079/BJN19980108
de Vos WM, Hugenholtz J (2004) Engineering metabolic highways in Lactococci and other lactic acid bacteria. Trends Biotechnol 22(2):72–79. https://doi.org/10.1016/j.tibtech.2003.11.011
Sendra E, Fayos P, Lario Y, Fernandez-Lopez J, Sayas-Barbera E, Perez-Alvarez JA (2008) Incorporation of citrus fibers in fermented milk containing probiotic bacteria. Food Microbiol 25(1):13–21. https://doi.org/10.1016/j.fm.2007.09.003
Majidzadeh Heravi R, Kermanshahi H, Sankian M, Nassiri MR, Moussavi AH, Nasiraii LR, Varasteh A (2016) Construction of a probiotic lactic acid bacterium that expresses acid resistant phytase enzyme. J Agric Sci Technol 18:925–936
Zuo RY, Chang JA, Yin QQ, Chen LY, Chen QX, Yang X, Zheng QH, Ren GZ, Feng H (2010) Phytase gene expression in Lactobacillus and analysis of its biochemical characteristics. Microbiol Res 165(4):329–335. https://doi.org/10.1016/j.micres.2009.06.001
Wang L, Yang YX, Cai B, Cao PH, Yang MM, Chen YL (2014) Coexpression and secretion of endoglucanase and phytase genes in Lactobacillus reuteri. Int J Mol Sci 15(7):12842–12860. https://doi.org/10.3390/ijms150712842
Igbasan FA, Manner K, Miksch G, Borriss R, Farouk A, Simon O (2000) Comparative studies on the in vitro properties of phytases from various microbial origins. Arch Tierernahr 53(4):353–373. https://doi.org/10.1080/17450390009381958
Pakbaten B, Majidzadeh Heravi R, Kermanshahi H, Sekhavati H, A J (2016) Construction of pFUM003 expression vector with extracellular secretion properties. Koomesh 17 (4):1017–1023
Mason CK, Collins MA, Thompson K (2005) Modified electroporation protocol for lactobacilli isolated frorn the chicken crop facilitates transformation and the use of a genetic tool. J Microbiol Methods 60(3):353–363. https://doi.org/10.1016/j.mimet.2004.10.013
Yin QQ, Zheng QH, Kang XT (2007) Biochemical characteristics of phytases from fungi and the transformed microorganism. Anim Feed Sci Technol 132:341–350. https://doi.org/10.1016/j.anifeedsci.2006.03.016
Han YM, Yang F, Zhou AG, Miller ER, Ku PK, Hogberg MG, Lei XG (1997) Supplemental phytases of microbial and cereal sources improve dietary phytate phosphorus utilization by pigs from weaning through finishing. J Anim Sci 75(4):1017–1025. https://doi.org/10.2527/1997.7541017x
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 227:2079–2085. https://doi.org/10.1038/227680a0
Bae HD, Yanke LJ, Cheng KJ, Selinger LB (1999) A novel staining method for detecting phytase activity. J Microbiol Methods 39:17–22. https://doi.org/10.1016/S0167-7012(99)00096-2
Gao Y, Shang C, Saghai Maroof MA, Biyashev RM, Grabau EA, Kwanyuen P, Burton JW, Buss GR (2007) A modified colorimetric method for Phytic acid analysis in soybean. Crop Sci 47:1797–1803. https://doi.org/10.2135/cropsci2007.03.0122
SASInstitute, (2004) SAS/STAT User’s guide: statistics. Version 9.2 Edition. SAS Inst. Inc., Cary, NC
Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi SH, Couloux A, Lee SW, Yoon SH, Cattolico L, Hur CG, Park HS, Segurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF (2009) Genome sequences of Escherichia coli B strains REL606 and BL21(DE3). J Mol Biol 394(4):644–652. https://doi.org/10.1016/j.jmb.2009.09.052
van Hartingsveldt W, van Zeijl CM, Harteveld GM, Gouka RJ, Suykerbuyk ME, Luiten RG, van Paridon PA, Selten GC, Veenstra AE, van Gorcom RF et al (1993) Cloning, characterization and overexpression of the phytase-encoding gene (phyA) of Aspergillus niger. Gene 127(1):87–94. https://doi.org/10.1016/0378-1119(93)90620-I
Borerro J, Jimenez j (2011) Use of the usp45 lactococcal secretion signal sequence to drive the secretion and functional expression of enterococcal bacteriocins in Lactococcus lactis. Appl Microbiol Biotechnol 89:131–143. https://doi.org/10.1007/s00253-010-2849-z
Neef J, Milder FJ, Koedijk DG, Klaassens M, Heezius EC, van Strijp JA, Otto A, Becher D, van Dijl JM, Buist G (2015) Versatile vector suite for the extracytoplasmic production and purification of heterologous his-tagged proteins in Lactococcus lactis. Appl Microbiol Biotechnol 99(21):9037–9048. https://doi.org/10.1007/s00253-015-6778-8
van Asseldonk M, Rutten G, Oteman M, Siezen RJ, de Vos WM, Simons G (1990) Cloning of usp45, a gene encoding a secreted protein from Lactococcus lactis subsp. lactis MG1363. Gene 95(1):155–160. https://doi.org/10.1016/0378-1119(90)90428-T
Le Loir Y, Nouaille S, Commissaire J, Bretigny L, Gruss A, Langella P (2001) Signal peptide and propeptide optimization for heterologous protein secretion in Lactococcus lactis. Appl Environ Microb 67(9):4119–4127. https://doi.org/10.1128/AEM.67.9.4119-4127.2001
Le Loir Y, Azevedo V, Oliveira SC, Freitas DA, Miyoshi A, Bermudez-Humaran LG, Nouaille S, Ribeiro LA, Leclercq S, Gabriel JE, Guimaraes VD, Oliveira MN, Charlier C, Gautier M, Langella P (2005) Protein secretion in Lactococcus lactis: an efficient way to increase the overall heterologous protein production. Microbial Cell Fact 4(1):2. https://doi.org/10.1186/1475-2859-4-2
Golovan S, Wang G, Zhang J, Forsberg CW (2000) Characterization and overproduction of the Escherichia coli appA encoded bifunctional enzyme that exhibits both phytase and acid phosphatase activities. Can J Microbiol 46(1):59–71. https://doi.org/10.1139/w99-084
Rodriguez E, Han Y, Lei XG (1999) Cloning, sequencing, and expression of an Escherichia coli acid phosphatase/phytase gene (appA2) isolated from pig colon. Biochem Biophys Res Commun 257(1):117–123. https://doi.org/10.1006/bbrc.1999.0361
Miksch G, Kleist S, Friehs K, Flaschel E (2002) Overexpression of the phytase from Escherichia coli and its extracellular production in bioreactors. Appl Microbiol Biotechnol 59(6):685–694. https://doi.org/10.1007/s00253-002-1071-z
Lee S, Kim T, Stahl CH, Lei XG (2005) Expression of Escherichia coli appA2 phytase in four yeast systems. Biotechnol Lett 27(5):327–334. https://doi.org/10.1007/s10529-005-0704-6
Ptak A, Józefiak D, Kierończyk B, Rawski M, Żyła K, Świątkiewicz S (2013) Effect of different phytases on the performance, nutrient retention and tibia composition in broiler chickens. Arch Tierernahr 56(104):1028–1038. https://doi.org/10.7482/0003-9438-56-104
Ravindran V, Morel PC, Partridge GG, Hruby M, Sands JS (2006) Influence of an Escherichia coli-derived phytase on nutrient utilization in broiler starters fed diets containing varying concentrations of phytic acid. Poult Sci 85(1):82–89. https://doi.org/10.1093/ps/85.1.82
De Angelis M, Gallo G, Corbo MR, McSweeney PL, Faccia M, Giovine M, Gobbetti M (2003) Phytase activity in sourdough lactic acid bacteria: purification and characterization of a phytase from Lactobacillus sanfranciscensis CB1. Int J Food Microbiol 87(3):259–270. https://doi.org/10.1016/S0168-1605(03)00072-2
Khodaii Z, Mehrabani Natanzi M, Naseri MH, Goudarzvand M, Dodson H, Snelling AM (2013) Phytase activity of lactic acid bacteria isolated from dairy and pharmaceutical probiotic products. Int J Enteric Pathog 1(1):12–16. https://doi.org/10.17795/ijep9359
Zamudio M, Gonzalez A, Medina JA (2001) Lactobacillus plantarum phytase activity is due to non-specific acid phosphatase. Lett Appl Microbiol 32(3):181–184. https://doi.org/10.1046/j.1472-765x.2001.00890.x
Greiner R, Konietzny U, Jany KD (1993) Purification and characterization of two phytases from Escherichia coli. Arch Biochem Biophys 303(1):107–113. https://doi.org/10.1006/abbi.1993.1261
Jiang XR, Luo F, Wu SG, Zhang HJ, Bontempo V, Qu RM, Yue HY, Qi GH (2015) Effect of phytase supplementation on apparent phosphorus digestibility and phosphorus output in broiler chicks fed low-phosphorus diets. Int J Health, Anim sci Food saf 1:9–17. https://doi.org/10.13130/2283-3927/4649
Acknowledgments
The authors gratefully acknowledge the financial support from the Excellence Centre for Animal Sciences, and Faculty of Agriculture, Ferdowsi University of Mashhad and Dr. Banayan Aval for his English editing service.
Funding
This study was funded by Ferdowsi university of Mashhad (FUM) 3/30173; 2/16/2014.
Author information
Authors and Affiliations
Contributions
BP carried out experimental process. RM designed and drafted the manuscript. HK and MS analyzed data and information of study. AJ conducted the study in laboratory. MM carried out animal model part of study. All author contributed in interpretation of results and read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Rights and permissions
About this article
Cite this article
Pakbaten, B., Majidzadeh Heravi, R., Kermanshahi, H. et al. Production of Phytase Enzyme by a Bioengineered Probiotic for Degrading of Phytate Phosphorus in the Digestive Tract of Poultry. Probiotics & Antimicro. Prot. 11, 580–587 (2019). https://doi.org/10.1007/s12602-018-9423-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12602-018-9423-x