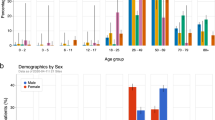
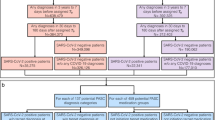
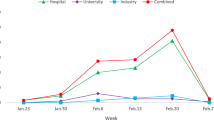
Abstract
For an emerging infectious disease such as 2019 coronavirus disease (COVID-19), initially there may not be any existing medication or treatment immediately available, which may result in high morbidity and mortality in a short time of period. In this case, it is urgent to quickly identify whether existing medications or treatments could be repurposed to treat the newly appeared disease before time-consuming randomized clinical trials (RCTs) can be done and new drugs can be developed. For example, when SARS-CoV-2 appeared in late 2019, clinicians started to use existing antiviral drugs, anti-inflammatory drugs, immune-based therapies and other types of medications to treat COVID-19 patients before any data or evidence was available to support the use of these medications for the new COVID-19 disease. Most of these medications have proven to be ineffective or only marginally effective to treat COVID-19 patients by more rigorous RCT or secondary data analyses later. We propose to use real-world electronic medical records (EMR) data to develop real-time treatment evaluation and monitoring systems to identify effective treatments or avoid ineffective treatments for emerging diseases in the future. In order to do this, first we have to deal with the challenges in processing and analyzing complex and noisy EMR data. In this paper, we outline these challenges and propose practical statistical methods and guidelines, which are derived from a project in evaluating anti-viral medication, remdesivir, for COVID-19 treatment based on a local healthcare EMR database.



Similar content being viewed by others
References
Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus, I., & Research, T (2020) A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382(8):727–733. https://doi.org/10.1056/NEJMoa2001017
Geneva: World Health Organization (2023) WHO COVID-19 Dashboard. https://covid19.who.int/
Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR et al (2020) Remdesivir for the treatment of Covid-19—final report. N Engl J Med 383(19):1813–1826. https://doi.org/10.1056/NEJMoa2007764
Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn M-Y, Nahass RG, Chen Y-S, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wei X, Gaggar A, Brainard DM et al (2020) Remdesivir for 5 or 10 days in patients with severe covid-19. N Engl J Med 383(19):1827–1837. https://doi.org/10.1056/NEJMoa2015301
Spinner CD, Gottlieb RL, Criner GJ, Arribas López JR, Cattelan AM, Soriano Viladomiu A, Ogbuagu O, Malhotra P, Mullane KM, Castagna A, Chai LYA, Roestenberg M, Tsang OTY, Bernasconi E, Le Turnier P, Chang S-C, SenGupta D, Hyland RH, Osinusi AO et al (2020) Effect of Remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA 324(11):1048–1057. https://doi.org/10.1001/jama.2020.16349
WHO Solidarity Trial Consortium (2020) Repurposed antiviral drugs for Covid-19—interim WHO solidarity trial results. N Engl J Med 384(6):497–511. https://doi.org/10.1056/NEJMoa2023184
Consortium, W. H. O. S. T. (2022) Remdesivir and three other drugs for hospitalised patients with COVID-19: final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet 399(10339):1941–1953. https://doi.org/10.1016/S0140-6736(22)00519-0
Thadhani R (2006) In: Mehta A, Beck M, Sunder-Plassmann G (eds) Fabry disease: perspectives from 5 years of FOS. https://www.ncbi.nlm.nih.gov/pubmed/21290683
Girman CJ, Ritchey ME, 3rd Lo Re V (2022) Real-world data: Assessing electronic health records and medical claims data to support regulatory decision-making for drug and biological products. Pharmacoepidemiol Drug Saf 31(7):717–720. https://doi.org/10.1002/pds.5444
Committee on the Learning Health Care System in, A., & Institute of, M. (2013). In M. Smith, R. Saunders, L. Stuckhardt, & J. M. McGinnis (Eds.), Best Care at Lower Cost: The Path to Continuously Learning Health Care in America. National Academies Press (US) Copyright 2013 by the National Academy of Sciences. All rights reserved. https://doi.org/10.17226/13444
Mills EJ, Thorlund K, Ioannidis JP (2013) Demystifying trial networks and network meta-analysis. BMJ 346:f2914. https://doi.org/10.1136/bmj.f2914
Tricoci P, Allen JM, Kramer JM, Califf RM, Smith SC Jr (2009) Scientific evidence underlying the ACC/AHA clinical practice guidelines. JAMA 301(8):831–841. https://doi.org/10.1001/jama.2009.205
Van Poucke S, Thomeer M, Heath J, Vukicevic M (2016) Are randomized controlled trials the (G)old standard? From clinical intelligence to prescriptive analytics. J Med Int Res 18(7):e185. https://doi.org/10.2196/jmir.5549
Anglemyer A, Horvath HT, Bero L (2014) Healthcare outcomes assessed with observational study designs compared with those assessed in randomized trials. Cochrane Database Syst Rev 2014(4):Mr000034. https://doi.org/10.1002/14651858.MR000034.pub2
Ioannidis JP, Haidich AB, Pappa M, Pantazis N, Kokori SI, Tektonidou MG, Contopoulos-Ioannidis DG, Lau J (2001) Comparison of evidence of treatment effects in randomized and nonrandomized studies. JAMA 286(7):821–830. https://doi.org/10.1001/jama.286.7.821
Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong IC, Rijnbeek PR, van der Lei J, Pratt N, Norén GN, Li YC, Stang PE, Madigan D, Ryan PB (2015) Observational health data sciences and informatics (OHDSI): opportunities for observational researchers. Stud Health Technol Inform 216:574–578
Hernán MA, Robins JM (2016) Using big data to emulate a target trial when a randomized trial is not available. Am J Epidemiol 183(8):758–764. https://doi.org/10.1093/aje/kwv254
Van Spall HG, Toren A, Kiss A, Fowler RA (2007) Eligibility criteria of randomized controlled trials published in high-impact general medical journals: a systematic sampling review. JAMA 297(11):1233–1240. https://doi.org/10.1001/jama.297.11.1233
Callahan A, Shah NH, Chen JH (2020) Research and reporting considerations for observational studies using electronic health record data. Ann Intern Med 172(11 Suppl):S79-s84. https://doi.org/10.7326/m19-0873
Kundu MG (2021) Statistics and machine learning methods for EHR data—from data extraction to data analytics. J Biopharm Stat 31(4):559–560. https://doi.org/10.1080/10543406.2021.1928833
Kratochwill TR, Bergan JR (1990) Treatment evaluation. In: TR Kratochwill, JR Bergan (eds) Behavioral consultation in applied settings: an individual guide. Springer, Berlin, pp 157–185. https://doi.org/10.1007/978-1-4757-9395-6_5
Shivade C, Raghavan P, Fosler-Lussier E, Embi PJ, Elhadad N, Johnson SB, Lai AM (2014) A review of approaches to identifying patient phenotype cohorts using electronic health records. J Am Med Inform Assoc 21(2):221–230. https://doi.org/10.1136/amiajnl-2013-001935
Zeng Z, Deng Y, Li X, Naumann T, Luo Y (2019) Natural language processing for EHR-based computational phenotyping. IEEE/ACM Trans Comput Biol Bioinform 16(1):139–153. https://doi.org/10.1109/tcbb.2018.2849968
Wu H, Yamal JM, Yaseen Y, Maroufy V (2021) Statistics and machine learning methods for EHR data: from data extraction to data analytics (edited). CRC Press, Boca Raton
Grubbs FE (1969) Procedures for detecting outlying observations in samples. Technometrics 11(1):1–21
Aguinis H, Gottfredson RK, Joo H (2013) Best-practice recommendations for defining, identifying, and handling outliers. Organ Res Methods 16(2):270–301
Barnett V, Lewis T (1994) Outliers in statistical data, 3rd edn. Wiley, New York
Box GEP, Cox DR (1964) An analysis of transformations. J R Stat Soc: Ser B (Methodol) 26(2):211–243. https://doi.org/10.1111/j.2517-6161.1964.tb00553.x
Dong Y, Peng CY (2013) Principled missing data methods for researchers. Springerplus 2(1):222. https://doi.org/10.1186/2193-1801-2-222
Graham JW (2009) Missing data analysis: making it work in the real world. Annu Rev Psychol 60:549–576. https://doi.org/10.1146/annurev.psych.58.110405.085530
Lachin JM (2016) Fallacies of last observation carried forward analyses. Clin Trials 13(2):161–168. https://doi.org/10.1177/1740774515602688
Jonsson P, Wohlin C (2004) An evaluation of k-nearest neighbour imputation using Likert data. In: 10th international symposium on software metrics, 2004. Proceedings
van Buuren S, Groothuis-Oudshoorn K (2011) mice: multivariate imputation by chained equations in R. J Stat Softw 45(3):1–67. https://doi.org/10.18637/jss.v045.i03
Azur MJ, Stuart EA, Frangakis C, Leaf PJ (2011) Multiple imputation by chained equations: what is it and how does it work? Int J Methods Psychiatr Res 20(1):40–49. https://doi.org/10.1002/mpr.329
Shah AD, Bartlett JW, Carpenter J, Nicholas O, Hemingway H (2014) Comparison of random forest and parametric imputation models. Am J Epidemiol 179(6):764–774
Molenberghs G, Kenward MG (2007) Missing data in clinical studies. Wiley, San Francisco, CA
Rai SN, Wu X, Srivastava DK, Craycroft JA, Rai JP, Srivastava S, James RF, Boakye M, Bhatnagar A, Baumgartner R (2020) Review: propensity score methods with application to the HELP clinic clinical study [Clinical report]. Open Access Medical Statistics, 11+. https://link.gale.com/apps/doc/A621084577/AONE?u=anon~b6653b08&sid=googleScholar&xid=666f20f0
Rosenbaum PR, Rubin DB (1983) The central role of the propensity score in observational studies for causal effects. Biometrika 70(1):41–55. https://doi.org/10.1093/biomet/70.1.41
Austin PC (2007) Propensity-score matching in the cardiovascular surgery literature from 2004 to 2006: a systematic review and suggestions for improvement. J Thorac Cardiovasc Surg 134(5):1128–1135. https://doi.org/10.1016/j.jtcvs.2007.07.021
Rosenbaum PR (1987) Model-based direct adjustment. J Am Stat Assoc 82(398):387–394. https://doi.org/10.2307/2289440
Kurita T (2019) Principal component analysis (PCA). In: Computer vision: a reference guide. Springer, Berlin, pp 1–4. https://doi.org/10.1007/978-3-030-03243-2_649-1
Fan J, Lv J (2008) Sure independence screening for ultrahigh dimensional feature space. J R Stat Soc: Ser B (Stat Methodol) 70(5):849–911. https://doi.org/10.1111/j.1467-9868.2008.00674.x
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc Ser B (Stat Methodol) 67(2):301–320. http://www.jstor.org/stable/3647580
Fan J, Li R (2001) Variable selection via nonconcave penalized likelihood and its oracle properties. J Am Stat Assoc 96(456):1348–1360. https://doi.org/10.1198/016214501753382273
Heinze G, Wallisch C, Dunkler D (2018) Variable selection—a review and recommendations for the practicing statistician. Biom J 60(3):431–449. https://doi.org/10.1002/bimj.201700067
Kaufman S, Rosset S, Perlich C (2011) Leakage in data mining: formulation, detection, and avoidance. In: Proceedings of the 17th ACM SIGKDD international conference on knowledge discovery and data mining, San Diego, California, USA. https://doi.org/10.1145/2020408.2020496
Chen Y, Wang J, Chubak J, Hubbard RA (2019) Inflation of type I error rates due to differential misclassification in EHR-derived outcomes: empirical illustration using breast cancer recurrence. Pharmacoepidemiol Drug Saf 28(2):264–268. https://doi.org/10.1002/pds.4680
Brown SM, Duggal A, Hou PC, Tidswell M, Khan A, Exline M, Park PK, Schoenfeld DA, Liu M, Grissom CK, Moss M, Rice TW, Hough CL, Rivers E, Thompson BT, Brower RG (2017) Nonlinear imputation of PaO2/FIO2 from SpO2/FIO2 among mechanically ventilated patients in the ICU: a prospective. Obs Study Crit Care Med 45(8):1317–1324. https://doi.org/10.1097/ccm.0000000000002514
Rosenbaum PR, Rubin DB (1985) Constructing a control group using multivariate matched sampling methods that incorporate the propensity score. Am Stat 39(1):33–38. https://doi.org/10.2307/2683903
Groenwold RHH (2020) Informative missingness in electronic health record systems: the curse of knowing. Diagn Progn Res 4(1):8. https://doi.org/10.1186/s41512-020-00077-0
Stekhoven DJ, Bühlmann P (2011) MissForest—non-parametric missing value imputation for mixed-type data. Bioinformatics 28(1):112–118. https://doi.org/10.1093/bioinformatics/btr597
Zhang C, Maroufy V, Chen B, Wu H (2021) Missing data issues in EHR. In: Statistics and machine learning methods for EHR data, 1st ed, p 25
Gray RJ (1988) A class of K-sample tests for comparing the cumulative incidence of a competing risk. Ann Stat 16(3):1141–1154. http://www.jstor.org/stable/2241622
Austin PC, Lee DS, Fine JP (2016) Introduction to the analysis of survival data in the presence of competing risks. Circulation 133(6):601–609. https://doi.org/10.1161/CIRCULATIONAHA.115.017719
Cole SR, Hernán MA (2008) Constructing inverse probability weights for marginal structural models. Am J Epidemiol 168(6):656–664. https://doi.org/10.1093/aje/kwn164
McCaw ZR, Tian L, Vassy JL, Ritchie CS, Lee C-C, Kim DH, Wei L-J (2020) How to quantify and interpret treatment effects in comparative clinical studies of COVID-19. Ann Intern Med 173(8):632–637. https://doi.org/10.7326/M20-4044
Scholz FW, Stephens MA (1987) K-sample anderson-darling tests. J Am Stat Assoc 82(399):918–924. https://doi.org/10.2307/2288805
Tibshirani R (1996) Regression shrinkage and selection via the Lasso. J R Stat Soc Series B (Methodol) 58(1):267–288. http://www.jstor.org/stable/2346178
Aleissa MM, Silverman EA, Acosta LMP, Nutt CT, Richterman A, Marty FM (2020) New perspectives on antimicrobial agents: Remdesivir treatment for COVID-19. Antimicrob Agents Chemother 65(1):e01814–e01820. https://doi.org/10.1128/AAC.01814-20
Ansems K, Grundeis F, Dahms K, Mikolajewska A, Thieme V, Piechotta V, Metzendorf MI, Stegemann M, Benstoem C, Fichtner F (2021) Remdesivir for the treatment of COVID-19. Cochrane Database Syst Rev 8(8):Cd014962. https://doi.org/10.1002/14651858.Cd014962
Arnaud M, Bégaud B, Thurin N, Moore N, Pariente A, Salvo F (2017) Methods for safety signal detection in healthcare databases: a literature review. Expert Opin Drug Saf 16(6):721–732. https://doi.org/10.1080/14740338.2017.1325463
Sacks JJ, Harrold LR, Helmick CG, Gurwitz JH, Emani S, Yood RA (2005) Validation of a surveillance case definition for arthritis. J Rheumatol 32(2):340–347
Cutler JA, Sorlie PD, Wolz M, Thom T, Fields LE, Roccella EJ (2008) Trends in hypertension prevalence, awareness, treatment, and control rates in United States adults between 1988–1994 and 1999–2004. Hypertension 52(5):818–827. https://doi.org/10.1161/hypertensionaha.108.113357
Kohsaka S, Katada J, Saito K, Jenkins A, Li B, Mardekian J, Terayama Y (2020) Safety and effectiveness of non-vitamin K oral anticoagulants versus warfarin in real-world patients with non-valvular atrial fibrillation: a retrospective analysis of contemporary Japanese administrative claims data. Open Heart 7(1):e001232. https://doi.org/10.1136/openhrt-2019-001232
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors do not have any financial interests that are directly or indirectly related to the work submitted for publication.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, C., Nigo, M., Patel, S. et al. Use of Real-World EMR Data to Rapidly Evaluate Treatment Effects of Existing Drugs for Emerging Infectious Diseases: Remdesivir for COVID-19 Treatment as an Example. Stat Biosci (2024). https://doi.org/10.1007/s12561-023-09411-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12561-023-09411-8