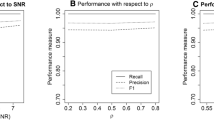
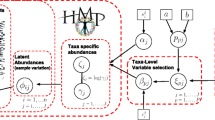
Abstract
The study of microbiome data has been widely used to investigate associations between the abundance of microbial taxa and human diseases. Identifying and understanding these relationships precisely gives the microbiome a key role in human health, disease status, and the development of new diagnostics and targeted therapeutics. Due to its unique features such as compositional data, excessive zero counts, overdispersion, and complexed structure between taxa, undertaking effective microbiome data analytics presents numerous obstacles. To quantify covariate-taxa effects on the subgingival microbiome study, we proposed a refined Bayesian zero-inflated negative binomial (ZINB) regression model with random subject effects. This proposed approach not only accommodates inflated zero counts and overdispersion similar to the existing ZINB model developed by Jiang et al. (Biostatistics 22(3):522–540, 2021), but also accounts for subject-level heterogeneity through the inclusion of random subject effects. In addition, an efficient Markov chain Monte Carlo (MCMC) sampling algorithm was developed for Bayesian computation. Overall effects of pre-selected group variables on predicted taxa abundance were estimated and tested under the proposed model. We conduct simulation studies and demonstrate that the proposed model outperforms the competing models in achieving a better power with controlling the type I error. The usefulness of the proposed model is applied to a real subgingival microbiome study.






Similar content being viewed by others
Code Availability
All code is written by a statistical software R 4.2.2 and the code can be available from the first author (yeongjin.gwon@unmc.edu).
References
Abraham C, Cho JH (2009) Inflammatory bowel disease. N Engl J Med 361:2066–2078
Ahn J, Sing R, Pei Z, Dominianni C, Wu J et al (2013) Human gut microbiome and risk for colorectal caner. J Natl Cancer Inst 105:1907–1911
Qin J, Li Y, Chi Z, Li S, Zhu J et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60
Romero R, Hassan SS, Gamer P, Barca AL, Faddish DW et al (2014) The vaginal microbiota of pregnant women who subsequently have spontaneous preterm labor and delivery and those with a normal delivery at term. Microbiome. https://doi.org/10.1186/2049-2618-2-18
Turnbaugh PJ, Ley RE, Mahowald MA, Margin V, Mardis ER, Gordon JI (2006) An obesity-associated gut microbiome with increases capacity for energy harvest. Nature 444(7122):1027–1031
Zhang X, Mallick H, Tang Z et al (2017) Negative binomial mixed models for analyzing microbiome count data. BMC Bioinform. https://doi.org/10.1186/s12859-016-1441-7
Chai H, Jiang H, Lin L, Liu L (2018) A marginalized two-part beta regression model for microbiome compositional data. Comput Biol 14(7):e1006329. https://doi.org/10.1371/journal.pcbi.1006329
Chen EZ, Li H (2016) A two-part mixed effects model for analyzing longitudinal microbiome compositional data. Bioinformatics 32:2611–2617
Ho NT, Li F, Wang S, Kuhn L (2019) metamicrobiomeR: an R package for analysis of microbiome relative abundance data using zero-inflated beta GAMLSS and meta-analysis across studies using random effects models. BMC Bioinform. https://doi.org/10.1186/s12859-019-2744-2
Peng X, Li G, Liu Z (2016) Zero-inflated beta regression for differential abundance analysis with metagenomics data. J Comput Biol 23:102–110
Tang Z, Chen G (2019) Zero-inflated generalized Dirichlet multinomial regression model for microbiome compositional data analysis. Biostatistics 20(4):698–713
Xia Y, Sun J, Chen DG (2018) Modeling zero-inflated microbiome data. In: Statistical analysis of microbiome data with R. ICSA book series in statistics. Springer, Singapore. https://doi.org/10.1007/978-981-13-1534-3_12
Jiang S, Xiao G, Koh AY, Kim J, Li Q, Zhang X (2021) A Bayesian zero-inflated negative binomial regression model for the integrative analysis of microbiome data. Biostatistics 22(3):522–540
Bakhshi E, Yazadanipour MA, Rahozar M et al (2019) Overall effects of risk factors associated with dental caries indices using the marginalized zero-inflated negative binomial model. Caries Res 53:541–546
Preisser JS, Das K, Long DL et al (2016) Marginalized zero-inflated negative binomial regression with application to dental caries. Stat Med 35:1722–1735
Smith VA, Neelon B, Preisser JS et al (2017) A marginalized two-part model for longitudinal semicontinuous data. Stat Methods Med Res 26:1949–1968
Zhang X, Yi N (2022) Analyzing the overall effects of the microbiome abundance data with a Bayesian predictive value approach. Stat Methods Med Res 31(10):1992–2003
Polson NG, Scott JG, Windle J (2013) Bayesian inference for logistic models using Polya-gamma latent variables. J Am Stat Assoc 108(504):1339–1349
Ibrahim JG, Chen M-H, Sinha D (2001) Bayesian survival analysis. Springer, New York
Mikuls TR, Walker C, Qiu F, Yu F, Thiele GM et al (2018) The sub gingival microbiome in patients with established rheumatoid arthritis. Rheumatology 57:1162–1172
Mikuls TR, Payne JB, Yu F, Thiele GM, Reynolds RJ et al (2014) Periodontitis and porphyromonas gingivitis in patients with rheumatoid arthritis. Arthritis Rheumatol 66:1090–1100
Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS et al (1988) The American rheumatism association 1987 revised criteria for the classification of rheumatoid arthritis. Arithritis Rheumatol 31(3):315–324
Qin H, Li G, Xu X, Zhang C, Zhong W, Xu S, Yin Y, Song J (2022) The role of oral microbiome in periodontitis under diabetes mellitus. J Oral Microbiol. https://doi.org/10.1080/20002297.2022.2078031
Zhou M, Carlin L (2015) Negative binomial process count and mixture modeling. IEEE Trans Pattern Anal Mach Intell 37:307–320
Geisser S, Eddy WF (1979) A predictive approach to model selection. J Am Stat Assoc 74:153–160
Zhang D, Chen M-H, Ibrahim JG, Boye ME, Shen W (2017) Bayesian model assessment in joint modeling of longitudinal and survival data with applications to cancer clinical trials. J Comput Graph Stat 26(1):121–133
Akherati M, Shafaei E, Salehiniya H, Abbaszadeh H (2021) Comparison of the frequency of periodontal pathogenic species of diabetics and non-diabetics and its relation to periodontitis severity, glycemic control and body mass index. Clin Exp Dent Res 7:1080–1088
Bourgeois D, Inquimbert C, Ottolenghi L, Carrouel F (2019) Periodontal pathogens as risk factors of cardiovascular diseases, diabetes, rheumatoid arthritis, cancer, and chronic obstructive pulmonary disease—is there cause for consideration? Microorganisms. https://doi.org/10.3390/microorganisms7100424
Omori M, Kato-Kogoe N, Sakaguchi S, Kamiya K, Fukui N, Gu YH et al (2021) Characterization of salivary microbiota in elderly patients with type 2 diabetes mellitus: a matched case–control study. Clin Oral Investig 26:493–504
Higgins JPT, Thomson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21(11):1539–1558
Chen M-H, Shao Q-M (1999) Monte Carlo estimation of Bayesian credible and HPD intervals. J Comput Graph Stat 8:69–92
Acknowledgements
We would like to thank the Editor, the Associate Editor, and the two anonymous reviewers for their very helpful and constructive comments along with suggestions. This has led to a substantial improved version of the article. This research was partially supported by a Veterans Affairs Merit Award [Grant Number CX000896] and the National Institute of General Medical Sciences (NIGMS) Grant #5U54GM115458. Dr. Payne’s research is supported by the F. Gene and Rosemary Dixon Endowed Chair in Dentistry.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 (pdf 930 KB)
Supplementary Information Online Supplementary Materials include six sections: (i) S1 (Summary of Taxa Information); (ii) S2 (Proof of Proposition 1); (iii) S3 (Derivation of Full Conditional Distributions); (iv) S4 (Additional Simulation Study based on Zero-Inflated Poisson Setting); (v) S5 (Additional Figures from Analysis of Subgingival Microbiome Study); and (vi) S6 (Trace and Autocorrelation Plots for the Selected Taxa from Analysis of Subgingival Microbiome Study). These will be available at the journal online website.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gwon, Y., Yu, F., Payne, J.B. et al. Bayesian Modeling on Microbiome Data Analysis: Application to Subgingival Microbiome Study. Stat Biosci (2023). https://doi.org/10.1007/s12561-023-09397-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12561-023-09397-3