Abstract
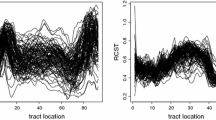
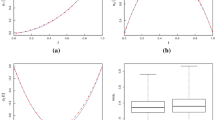
Functional data are often generated by modern biomedical technologies where features related to the pathophysiology and pathogenesis of a disease are interrogated repeatedly over time and at multiple spatially interdependent regions. To reduce model complexity and simplify the resulting inference, possible spatial correlation among neighboring regions is often neglected. In this article, we propose a weighted kernel smoothing estimate of the mean function that leverages the spatial and temporal correlation. We also address the companion problem of developing a simultaneous prediction method for individual curves using discrete samples. We establish the asymptotic properties of the proposed estimate, including its unique maximum efficiency achieving minimum asymptotic variance. The proposed method improves estimation and prediction in the presence of sparse observations, and therefore, is advantageous to biomedical applications that utilize markers to identify features intrinsic to a particular disease at multiple interdependent sites within an organ. Our simulation and case studies show that the proposed method outperforms conventional approaches for characterizing the dynamic functional imaging data, with the maximum benefit achieved in the presence of a small number of repeated scans.




Similar content being viewed by others
References
Baladandayuthapani V, Mallick BK, Hong MY, Lupton JR, Turner ND, Carroll RJ (2008) Bayesian hierarchical spatially correlated functional data analysis with application to colon carcinogenesis. Biometrics 64:64–73
Chen K, Müller H (2012) Modeling repeated functional observations. JASA 107(500):1599–1609
Degras D (2011) Simultaneous confidence bands for nonparametric regression with functional. Statistica Sinica 21:1735–1765
Gelman A, Imbens G (2014) Why high-order polynomials should not be used in regression discontinuity designs NBER Working Paper No. 20405
Goldsmith J, Greven S, Crainiceanu C (2012) Corrected confidence bands for functional data using principal components. Biometrics 69:41–51
Guo W (2002) Functional mixed effect model. Biometrics 58:121–128
Lee T-Y (2002) Functional CT: physiological models. Trends Biotechnol 20(8 Suppl):S3–S10
Li Y, Wang N, Hong M, Turner ND, Lupton JR, Carroll RJ (2007) Nonparametric estimation of correlation functions in longitudinal and spatial data, with application to colon carcinogenesis experiments. Ann Stat 35:1608–1643
Lin X, Carroll RJ (2000) Nonparametric function estimation for clustered data when the predictor is measured without/with error. J Am Stat Assoc 95:520–534
Miles K (2002) Functional computed tomography in oncology. Eur J Cancer 38:2079–2084
Miles K, Charnsangavej C, Lee F, Fishman E, Horton K, Lee T-Y (2000) Application of CT in the investigation of angiogenesis in oncology. Acad Radiol 7:840–850
Morris JS, Carroll RJ (2006) Wavelet-based functional mixed models. J R Stat Soc B 68:179–199
Ng C, Hobbs B, Chandler A, Anderson E, Herron D, Charnsangavej C, Yao J (2013) Metastases to the liver from neuroendocrine tumors: effect of duration of scan acquisition on CT perfusion values. Radiology 269:758–767
Park SY, Staicu AM (2015) Longitudinal functional data analysis. STAT 4:212–226
Paul D, Peng J (2011) Principal components analysis for sparsely observed correlated functional data using a kernel smoothing approach. Electron J Stat 5:1960–2003
Ramsay JO, Silverman BW (2005) Functional data analysis. Springer, New York
Rice J, Silverman B (1991) Estimating the mean and covariance structure nonparametrically when the data are curves. J R Stat Soc Ser B 53:233–243
Staicu A-M, Crainiceanu CM, Carroll RJ (2010) Fast methods for spatially correlated multilevel functional data. Biostatistics 11:177–194
Stewart EE, Chen X, Hadway J, Lee T (2008) Hepatic perfusion in a tumor model using DCE-CT: an accuracy and precision study. Phys Med Biol 53:4249–4267
Wang Y, Hobbs BP, Hu J, Ng C, Do K (2015) Predictive classification of correlated targets with application to detection of metastatic cancer using functional CT imaging. Biometrics 71:792–802
Yao F, Müller H-G, Wang J-L (2005) Functional data analysis for sparse longitudinal data. J Am Stat Assoc 100:577–590
Zhang L, Baladandayuthapani V, Zhu H, Baggerly KA, Majewski T, Czerniak B, Morris JS (2015) Functional car models for spatially correlated functional datasets. J Am Stat Assoc 111:772–786
Zhou L, Huang JZ, Carroll RJ (2008) Joint modeling of paired sparse functional data using principal components. Biometrika 95:601–619
Zhou L, Huang JZ, Martinez JG, Maity A, Baladandayuthapani V, Carroll RJ (2010) Reduced rank mixed effects models for spatially correlated hierarchical functional data. J Am Stat Assoc 105:390–400
Acknowledgements
This research was partially supported by NIH/NCI 5 P30 CA013696 Columbia University CCSG. KAD is partially supported by NIH/NCI CCSG Grant P30CA016672, EDRN Grant U24CA086368-19, SPORE Grant P50CA140388-08, CCTS Grant 5UL1TR000371-07, CPRIT Grants RP150006 and RP160693, MD Anderson Moonshot programs 710499-80-111529-19, 710499-80-116550-21, 710499-80-111995-21.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Materials:
The online supplementary materials include the proofs for Theorem 1-4, implementation of the hierarchical model in Baladandayuthapani et al. [1], empirical investigation for consistency of the spatial correlation parameter and noise variance. (pdf 270KB)
Rights and permissions
About this article
Cite this article
Wang, Y., Hu, J., Do, KA. et al. An Efficient Nonparametric Estimate for Spatially Correlated Functional Data. Stat Biosci 11, 162–183 (2019). https://doi.org/10.1007/s12561-019-09233-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12561-019-09233-7