Abstract
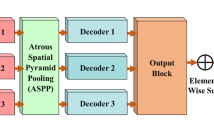
Automatic multimodal image segmentation is considered a challenging research area in the biomedical field. U-shaped models have led to an enormous breakthrough in a large domain of medical image segmentation in recentyears. The receptive field plays an essential role in convolutionalneural networks because too small a receptive field limits context information, and too large loses localization accuracy. Despite outstanding overall performance in biomedical segmenting, classical UNet architecture uses a fixed receptive field in convolutions operations. This study proposes a few modifications in classical UNet architecture by adjusting the receptive field via feature-fused module and attention gate mechanism. Compared with baseline UNet, the numerical parameters of FF-UNet (3.94 million) is 51% of classical UNet architecture (7.75 million). Furthermore, we extended our model performance by introducing post-processing schemes. The tri-threshold fuzzy intensification-based contrast enhancement technique is utilized to improve the contrast of biomedical datasets. In the second tier, the black top-hat filtering-based method is employed to remove hair-like artifacts from the ISIC 2018 skin lesion dataset, which may create a barrier to correctly segmenting the images. The proposed models have been trained using fivefold cross-validation on five publicly available biomedical datasets and achieved the dice coefficients of 0.860, 0.932, 0.932, 0.925, and 0.894 on ETIS-LaribPolypDB, CVC-ColonDB, CVC-ClinicDB, DSB 2018, and ISIC 2018 datasets, respectively. To further verify our claims, comparative analysis based on dice results is conducted, proving the proposed model effectiveness. The FF-UNet implementation models and pre-trained weights are freely publicly available: https://github.com/ahmedeqbal/FF-UNet.










Similar content being viewed by others
References
Tashk A, Herp J, Nadimi E. Fully automatic polyp detection based on a novel U-Net architecture and morphological post-process. Proc - 2019 3rd Int Conf Control Artif Intell Robot Optim ICCAIRO 2019. 2019;37–41.
American Cancer Society, Atlanta G. American Cancer Society: cancer facts &figures 2021. [Internet]. 2021. Available from: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/cancer-facts-figures-2021.html
Park CH, Kim JO, Choi MG, Kim KJ, Kim YH, Kim YS, et al. Utility of capsule endoscopy for the classification of Crohn’s disease: A multicenter study in Korea. Dig Dis Sci. 2007;52:1405–9.
Choi HN, Kim HH, Oh JS, Jang HS, Hwang HS, Kim EY, et al. Factors influencing the miss rate of polyps in a tandem colonoscopy study. Korean J Gastroenterol. 2014;64:24–30.
Rabeneck L, Souchek J, El-Serag HB. Survival of colorectal cancer patients hospitalized in the Veterans Affairs Health Care System. Am J Gastroenterol [Internet]. 2003;98:1186–92. Available from: https://journals.lww.com/00000434-200305000-00038
Rigel DS, Friedman RJ, Kopf AW. The incidence of malignant melanoma in the United States: issues as we approach the 21st century. J Am Acad Dermatol. 1996;34:839–47.
Khan MA, Akram T, Sharif M, Shahzad A, Aurangzeb K, Alhussein M, et al. An implementation of normal distribution based segmentation and entropy controlled features selection for skin lesion detection and classification. BMC Cancer BMC Cancer. 2018;18:1–20.
Barata C, Ruela M, Francisco M, Mendonca T, Marques JS. Two systems for the detection of melanomas in dermoscopy images using texture and color features. IEEE Syst J [Internet]. 2014;8:965–79. Available from: http://ieeexplore.ieee.org/document/6570764/
Losina E, Walensky RP, Geller A, Beddingfield FC, Wolf LL, Gilchrest BA, et al. Visual screening for malignant melanoma. Arch Dermatol [Internet]. 2007;143:21–8. Available from: http://archderm.jamanetwork.com/article.aspx?doi=10.1001/archderm.143.1.21
Caicedo JC, Goodman A, Karhohs KW, Cimini BA, Ackerman J, Haghighi M, et al. Nucleus segmentation across imaging experiments: the 2018 Data Science Bowl. Nat Methods [Internet]. Springer US; 2019;16:1247–53. Available from: https://doi.org/10.1038/s41592-019-0612-7
Bennai MT, Guessoum Z, Mazouzi S, Cormier S, Mezghiche M. A stochastic multi-agent approach for medical-image segmentation: application to tumor segmentation in brain MR images. Artif Intell Med. 2020;110.
Lundervold AS, Lundervold A. An overview of deep learning in medical imaging focusing on MRI. Z Med Phys [Internet]. Elsevier B.V.; 2019;29:102–27. Available from: https://doi.org/10.1016/j.zemedi.2018.11.002
De Brabandere B, Jia X, Tuytelaars T, Van Gool L. Dynamic filter networks. Adv Neural Inf Process Syst [Internet]. 2016;667–75. Available from: http://arxiv.org/abs/1605.09673
Qin X, Wu C, Chang H, Lu H, Zhang X. Match Feature U-Net: dynamic receptive field networks for biomedical image segmentation. symmetry (Basel) [Internet]. 2020;12:1230. Available from: https://www.mdpi.com/2073-8994/12/8/1230
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. 2015. p. 234–41. Available from: http://link.springer.com/10.1007/978-3-319-24574-4_28
Milletari F, Navab N, Ahmadi SA. V-Net: fully convolutional neural networks for volumetric medical image segmentation. Proc - 2016 4th Int Conf 3D Vision, 3DV 2016. 2016;565–71.
Ibtehaz N, Rahman MS. MultiResUNet: rethinking the U-Net architecture for multimodal biomedical image segmentation. Neural Networks [Internet]. Elsevier Ltd; 2020;121:74–87. Available from: https://doi.org/10.1016/j.neunet.2019.08.025
Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) [Internet]. 2015;9351:234–41. Available from: http://arxiv.org/abs/1505.04597
Jha D, Riegler MA, Johansen D, Halvorsen P, Johansen HD. DoubleU-Net: a deep convolutional neural network for medical image segmentation. Proc - IEEE Symp Comput Med Syst. 2020;2020-July:558–64.
Byra M, Jarosik P, Szubert A, Galperin M, Ojeda-Fournier H, Olson L, et al. Breast mass segmentation in ultrasound with selective kernel U-Net convolutional neural network. Biomed Signal Process Control [Internet]. 2020;61:102027. Available from: https://linkinghub.elsevier.com/retrieve/pii/S174680942030183X
Iqbal A, Sharif M. MDA-Net: Multiscale dual attention-based network for breast lesion segmentation using ultrasound images. J King Saud Univ - Comput Inf Sci [Internet]. 2021; Available from: https://linkinghub.elsevier.com/retrieve/pii/S1319157821002895
Cheng J, Tian S, Yu L, Lu H, Lv X. Fully convolutional attention network for biomedical image segmentation. Artif Intell Med [Internet]. Elsevier; 2020;107:101899. Available from: https://doi.org/10.1016/j.artmed.2020.101899
Hasan MK, Dahal L, Samarakoon PN, Tushar FI, Martí R. DSNet: Automatic dermoscopic skin lesion segmentation. Comput Biol Med [Internet]. Elsevier Ltd; 2020;120:103738. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0010482520301190
Mahmud T, Paul B, Fattah SA. PolypSegNet: A modified encoder-decoder architecture for automated polyp segmentation from colonoscopy images. Comput Biol Med [Internet]. Elsevier Ltd; 2021;128:104119. Available from: https://doi.org/10.1016/j.compbiomed.2020.104119
Jin Q, Cui H, Sun C, Meng Z, Su R. Cascade knowledge diffusion network for skin lesion diagnosis and segmentation. Appl Soft Comput [Internet]. Elsevier B.V.; 2021;99:106881. Available from: https://doi.org/10.1016/j.asoc.2020.106881
Nguyen NQ, Vo DM, Lee SW. Contour-aware polyp segmentation in colonoscopy images using detailed upsamling encoder-decoder networks. IEEE Access. 2020;8:99495–508.
Lei B, Xia Z, Jiang F, Jiang X, Ge Z, Xu Y, et al. Skin lesion segmentation via generative adversarial networks with dual discriminators. Med Image Anal [Internet]. Elsevier B.V.; 2020;64:101716. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1361841520300803
Qamar S, Ahmad P, Shen L. Dense encoder-decoder–based architecture for skin lesion segmentation. Cognit Comput [Internet]. 2021;13:583–94. Available from: http://link.springer.com/10.1007/s12559-020-09805-6
Silva J, Histace A, Romain O, Dray X, Granado B. Toward embedded detection of polyps in WCE images for early diagnosis of colorectal cancer. Int J Comput Assist Radiol Surg. 2014;9:283–93.
Bernal J, Sánchez J, Vilariño F. Towards automatic polyp detection with a polyp appearance model. Pattern Recognit. 2012;45:3166–82.
Bernal J, Sánchez FJ, Fernández-Esparrach G, Gil D, Rodríguez C, Vilariño F. WM-DOVA maps for accurate polyp highlighting in colonoscopy: validation vs saliency maps from physicians. Comput Med Imaging Graph. 2015;43:99–111.
Shorten C, Khoshgoftaar TM. A survey on image data augmentation for deep learning. J Big Data [Internet]. Springer International Publishing; 2019;6. Available from: https://doi.org/10.1186/s40537-019-0197-0
Al-Ameen Z. Visibility enhancement for images captured in dusty weather via tuned tri-threshold fuzzy intensification operators. Int J Intell Syst Appl. 2016;8:10–7.
Morphological image analysis: principles and applications. Sens Rev [Internet]. 2000;20. Available from: https://www.emerald.com/insight/content/doi/10.1108/sr.2000.08720cae.001/full/html
Li X, Wang W, Hu X, Yang J. Selective kernel networks. 2019 IEEE/CVF Conf Comput Vis Pattern Recognit [Internet]. IEEE; 2019. p. 510–9. Available from: https://ieeexplore.ieee.org/document/8954149/
Oktay O, Schlemper J, Folgoc L Le, Lee M, Heinrich M, Misawa K, et al. Attention U-Net: learning where to look for the pancreas. arXiv [Internet]. 2018; Available from: http://arxiv.org/abs/1804.03999
Schlemper J, Oktay O, Schaap M, Heinrich M, Kainz B, Glocker B, et al. Attention gated networks: learning to leverage salient regions in medical images. Med Image Anal [Internet]. 2019;53:197–207. Available from: https://linkinghub.elsevier.com/retrieve/pii/S1361841518306133
Shelhamer E, Long J, Darrell T. Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell [Internet]. 2017;39:640–51. Available from: http://ieeexplore.ieee.org/document/7478072/
Chen LC, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40:834–48.
Zhang L, Dolwani S, Ye X. Automated polyp segmentation in colonoscopy frames using fully convolutional neural network and textons. Commun Comput Inf Sci. 2017;723:707–17.
Akbari M, Mohrekesh M, Nasr-Esfahani E, Soroushmehr SMR, Karimi N, Samavi S, et al. Polyp segmentation in colonoscopy images using fully convolutional network. 2018 40th Annu Int Conf IEEE Eng Med Biol Soc [Internet]. IEEE; 2018. p. 69–72. Available from: https://ieeexplore.ieee.org/document/8512197/
Alom MZ, Yakopcic C, Hasan M, Taha TM, Asari VK. Recurrent residual U-Net for medical image segmentation. J Med Imaging [Internet]. 2019;6:1. Available from: https://www.spiedigitallibrary.org/journals/journal-of-medical-imaging/volume-6/issue-01/014006/Recurrent-residual-U-Net-for-medical-image-segmentation/10.1117/1.JMI.6.1.014006.full
Azad R, Asadi-Aghbolaghi M, Fathy M, Bi-directional ES, U-net ConvLSTM, with densley connected convolutions. Proc -,. Int Conf Comput Vis Work ICCVW 2019. IEEE. 2019;2019:406–15.
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imaging. 2020;39:1856–67.
Jha D, Smedsrud PH, Riegler MA, Johansen D, De Lange T, Halvorsen P, ResUNet++: an advanced architecture for medical image segmentation. Proc -, et al. IEEE Int Symp Multimedia. ISM. 2019;2019(2019):225–30.
Fujita S, Han XH. Cell detection and segmentation in microscopy images with improved mask R-CNN. Proc Asian Conf … [Internet]. 2020;1–13. Available from: https://openaccess.thecvf.com/content/ACCV2020W/MLCSA/papers/Fujita_Cell_Detection_and_Segmentation_in_Microscopy_Images_with_Improved_Mask_ACCVW_2020_paper.pdf
Natarajan VA, Sunil Kumar M, Patan R, Kallam S, Noor Mohamed MY. Segmentation of nuclei in histopathology images using fully convolutional deep neural architecture. 2020 Int Conf Comput Inf Technol ICCIT. 2020;1:319–25.
Banik D, Bhattacharjee D, Nasipuri M. A multi-scale patch-based deep learning system for polyp segmentation. Adv Intell Syst Comput. 2020;1136:109–19.
Olimov B, Sanjar K, Din S, Ahmad A, Paul A, Kim J. FU-Net: fast biomedical image segmentation model based on bottleneck convolution layers. Multimed Syst [Internet]. Springer Berlin Heidelberg; 2021;27:637–50. Available from: https://doi.org/10.1007/s00530-020-00726-w
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Ethical Approval
Not applicable.
Conflict of Interest
The authors declareno competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Iqbal, A., Sharif, M., Khan, M.A. et al. FF-UNet: a U-Shaped Deep Convolutional Neural Network for Multimodal Biomedical Image Segmentation. Cogn Comput 14, 1287–1302 (2022). https://doi.org/10.1007/s12559-022-10038-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12559-022-10038-y