Abstract
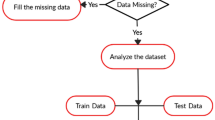
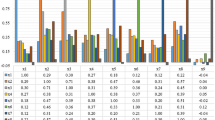
Cervical cancer is fourth main causes of death in women. Cervix is the main origin of cervical cancer. The idea of this research is to explore and propose an efficient and improved prediction method of cervical cancer. Earlier detection and prediction methods/test were very complex, tedious and requires medical and pathological expertise. In this paper, Machine learning approach is used for prediction and detection of cervical cancer. Integrated approach of Genetic Algorithm and Adaptive Boosting is used for performance evaluation for prediction of disease. Genetic algorithm is used as attribute selector to decrease the number of attributes. This not only declines the computational cost but also reduces the number of parameters for diagnosis. Adaptive Boosting is used to improve the performance of classifiers. C 4.5 Decision Tree and Support Vector Machine (SVM) are proposed for prediction of disease. Initially 32 attributes are used for prediction of cervical cancer. The numbers of attributes are reduced with genetic algorithm and further performance enhancement is proposed with adaptive boosting technique. With proposed integrated approach of genetic algorithm and adaptive boosting the improved accuracy lies between 94.17%-94.69%, sensitivity 97.36%-98.90%, specificity 93.37%-94.72% and precision 93%-95.17% for Support Vector Machine Radial Bias Function (SVM RBF), SVM Linear and Decision Tree.





Similar content being viewed by others
References
World Health Organization report on cervical cancer available at www.who.int
Exner M, Kuhn A, Stumpp P. Value of diffusion-weighted MRI in diagnosis of uterine cervical cancer: A prospective study evaluating the benefits of DWI compared to conventional MR sequences in a 3T environment. Acta Radiol. 2016;57(7):869–77.
Mcveigh PZ, Syed AM, Milosevic M, Fyles A, Haider MA. Diffusion-weighted MRI in cervical cancer. Eur Radiol. 2008;18(5):1058–64.
Duraisamy K, Jaganathan KS, Bose JC. Methods of Detecting Cervical Cancer. Adv Biol Res. 2011;5(4):226–32.
Brown AJ, Trimble CL. New Technologies for Cervical Cancer Screening. Best Pract Res Clin Obstet Gynaecol. 2012;26(2):233–42.
Raifu AO, El-Zein M, Sangwa-Lugoma G, Ramanakumar A, Walte SD. Determinants of Cervical Cancer Screening Accuracy for Visual Inspection with Acetic Acid (VIA) and Lugol's Iodine (VILI) Performed by Nurse and Physician. PLoS One. 2017;12(1):e0170631.
Gadducci A, Barsotti C, Cosio S, Domenici L, Riccardo AG. Smoking habit, immune suppression, oral contraceptive use, and hormone replacement therapy use and cervical carcinogenesis: A review of the literature. Gynecol Endocrinol. 2011;27(8):597–604.
Luhn P, Walker J, Schiffman M, Zuna RE. The role of co-factors in the progression from human papillomavirus infection to cervical cancer. Gynecol Oncol. 2013;128(2):265–70.
Cervical Cancer Prevention. Available: https://www.cancer.gov/types/cervical/hp/cervical-prevention-pdq. 2015.
Ronco G, Dillner J, Elfström KM, Tunesi S, Snijders PJ, Arbyn M, et al. Efficacy of HPV-based screening for prevention of invasive cervical cancer: follow-up of four European randomised controlled trials. Lancet. 2014;383:524–32.
Galgano MT, Castle PE, Atkins KA, Brix WK, Nassau SR, Stoler MH. Using biomarkers as objective standards in the diagnosis of cervical biopsies. Am J Surg Pathol. 2010;34:1077.
Ramaraju H, Nagaveni Y, Khazi A. Use of Schiller’s test versus Pap smear to increase detection rate of cervical dysplasias. International Journal of Reproduction, Contraception, Obstetrics and Gynecology. 2017;5:1446–50.
Kusy M, Obrzut B, Kluska J. Application of gene expression programming and neural networks to predict adverse events of radical hysterectomy in cervical cancer patients. Med Biol Eng Comput. 2013;51:1357–65.
Sara Moein. Medical Diagnosis using Neural Networks. IGI Global, 2014. 1-310. Web. 29 Aug. 2019. https://doi.org/10.4018/978-1-4666-6146-2.
Kononenko I. Machine learning for medical diagnosis: history, state of the art and perspective. Artif Intell Med. 2001;23:89–109.
Deo RC. Machine Learning in Medicine. Circulation. 2015;132(20):1920–30.
Fernandes K, Chicco D, Cardoso JS, Fernandes J. Supervised Deep Learning Embeddings for the Prediction of Cervical Cancer Diagnosis. PeerJ Computer Science. 2018;4:e154.
Fernandes K, Cardoso JS, Fernandes J. Transfer Learning With Partial Observability Applied to Cervical Cancer Screening. In: Alexandre L, Salvador Sańchez J, Rodrigues J, editors. Iberian, Conference on Pattern Recognition and Image Analysis. Faro: Springer; 2017. p. 243–50.
Wu W, Zhou H. Data-Driven Diagnosis of Cervical Cancer With Support Vector Machine-Based Approaches. IEEE Access. 2017;5:25189–95.
Salmeron JL, Rahimi SA, Navali AM, Sadeghpour A. Medical Diagnosis of Rheumatoid Arthritis Using Data Driven PSO-FCM With Scarce Datasets. Neurocomputing. Apr. 2017;232:104–12.
Jassim G, Obeid A, Nasheet HAA. Knowledge, Attitudes, And Practices Regarding Cervical Cancer And Screening Among Women Visiting Primary Health Care Centres In Bahrain. BMC Public Health. 20187;18, 2018(128). https://doi.org/10.1186/s12889-018-5023-7.
Singh A, Pandey B. A New Intelligent Medical Decision Support System Based on Enhanced Hierarchical Clustering and Random Decision Forest for the Classification of Alcoholic Liver Damage, Primary Hepatoma, Liver Cirrhosis, and Cholelithiasis. Journal of Healthcare Engineering. 2018;2018:1469043.
Hastie T, Tibshirani R, Friedman J. The Elements of Statistical Learning. New York: Springer Science & Business Media; 2009.
Abu-Mostafa YS, Magdon-Ismail M, Lin H-T. Learning from Data. 2012. AMLbook.com.
De Fauw J, et al. Clinically Applicable Deep Learning For Diagnosis and Referral in Retinal Disease. Nat Med. Sep 2018;24(9):1342–50.
Wu M, Yan C, Liu H, Liu Q. Automatic Classification of Ovarian Cancer Types From Cytological Images Using Deep Convolutional Neural Networks. Biosci Rep. 2018;38:BSR20180289. https://doi.org/10.1042/BSR20180289.
Liang X, Zhu L, Huang D-S. Multi-Task Ranking SVM For Image Cosegmentation. Neurocomputing. 2017;247:126–36.
Bolón-Canedo V, Ataer-Cansizoglu E, Erdogmus D, KalpathyCramer J, Fontenla-Romero O, Alonso-Betanzos A, et al. Dealing With Inter-Expert Variability in Retinopathy of Prematurity: A Machine Learning Approach. Comput Methods Prog Biomed. 2015;122(1):1–15.
Bolón-Canedo V, Remeseiro B, Alonso-Betanzos A, Campilho A. Machine Learning for Medical Applications, ESANN 2016 proceedings, European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning. Bruges (Belgium), 2016, pp 225-234.
Cortes C, Vapnik V. Support-Vector Network. Mach Learn. 1995;20:273–97.
Hsu C-W, Lin C-J. A Comparison of Methods for Multi-Class Support Vector Machines. IEEE Trans Neural Netw. 2002;13(2):415–25.
Joachims T (1998) Text categorization with support vector machines: learning with many relevant features. in Proceedings of ECML-98, 10th European Conference on Machine Learning, number 1398, pp. 137–142.
Quinlan JR. C4.5: Programs for Machine Learning, The Morgan Kaufmann series in machine learning, Elsevier, 2014.
He H, Garcia EA. Learning from imbalanced data. IEEE Trans Knowledge Data Eng. 2009;21(9):1263–84.
Daskalaki S, Kopanas I, Avouris N. Evaluation of classifiers for an uneven class distribution problem. Appl Artif Intell. 2006;20(5):381–417.
Blagus R, Lusa L. Class prediction for high-dimensional class-imbalanced data. BMC Bioinformatics. 2010;11:523.
Hulse JV, Khoshgoftaar TM, Napolitano A: Experimental perspectives on learning from imbalanced data. In Proceedings of the 24th international conference on Machine learning. Corvallis: Oregon State University; 2007:935–942.
Chawla NV, Bowyer KW, Hall LO, Philip Kegelmeyer W. SMOTE: Synthetic Minority Over-sampling Technique. J Artif Intell Res. 2002;16:321–57.
Cieslak DA, Chawla NW. Striegel A: Combating Imbalance in Network Intrusion Datasets. In Proc IEEE Int. Conf Granular Comput. Atlanta; 2006:732–737.
Fallahi A, Jafari S. An Expert System for Detection of Breast Cancer Using Data Pre processing and Bayesian Network. Int J AdvSci Technol. 2011;34:65–70.
Liu Y, Chawla NV, Harper MP, Shriberg E, Stolcke A. A Study In Machine Learning From Imbalanced Data For Sentence Boundary Detection In Speech. Comput Speech Lang. 2006;20(4):468–94.
MacIsaac KD, Gordon DB, Nekludova L, Odom DT, Schreiber J, Gifford DK, et al. A Hypothesis-Based Approach For Identifying The Binding Specificity of Regulatory Proteins From Chromatin Immuno precipitation Data. Bioinformatics. 2006;22(4):423–9.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sharma, M. Cervical cancer prognosis using genetic algorithm and adaptive boosting approach. Health Technol. 9, 877–886 (2019). https://doi.org/10.1007/s12553-019-00375-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12553-019-00375-8