Abstract
Intrinsically disordered proteins (IDPs) are proteins that lack rigid 3D structure but exist as conformational ensembles. Because of their structural plasticity, they can interact with multiple partners. The protein interactions between IDPs and their partners form scale-free protein interaction networks (PINs) that facilitate information flow in the cell. Because of their plasticity, IDPs typically occupy hub positions in cellular PINs. Furthermore, their conformational dynamics and propensity for post-translational modifications contribute to “conformational” noise which is distinct from the well-recognized transcriptional noise. Therefore, upregulation of IDPs in response to a specific input, such as stress, contributes to increased noise and, hence, an increase in stochastic, “promiscuous” interactions. These interactions lead to activation of latent pathways or can induce “rewiring” of the PIN to yield an optimal output underscoring the critical role of IDPs in regulating information flow. We have used PAGE4, a highly intrinsically disordered stress-response protein as a paradigm. Employing a variety of experimental and computational techniques, we have elucidated the role of PAGE4 in phenotypic switching of prostate cancer cells at a systems level. These cumulative studies over the past decade provide a conceptual framework to better understand how IDP conformational dynamics and conformational noise might facilitate cellular decision-making.

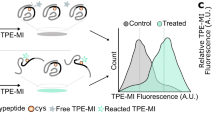
Reproduced with permission from Mahmoudabadi et al. 2013

Reproduced with permission from Rajagopalan et al. 2014

Reproduced with permission from Kulkarni et al. 2017


Reproduced with permission from Kulkarni et al. 2017

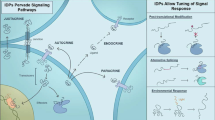
Reproduced with permission from Singh et al. Entropy (Basel). 2021 Feb 26;23(3):288
Similar content being viewed by others
Data availability
Not applicable.
Code availability
Not applicable.
References
Al Emran A, Marzese DM, Menon DR, Stark MS, Torrano J, Hammerlindl H, Zhang G, Brafford P, Salomon MP, Nelson N, Hammerlindl S, Gupta D, Mills GB, Lu Y, Sturm RA, Flaherty K, Hoon DSB, Gabrielli B, Herlyn M, Schaider H (2017) Distinct histone modifications denote early stress-induced drug tolerance in cancer. Oncotarget 9(9):8206–8222. https://doi.org/10.18632/oncotarget.23654
Arai M, Sugase K, Dyson HJ, Wright PE (2015) Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding. Proc Natl Acad Sci U S A 112(31):9614–9619. https://doi.org/10.1073/pnas.1512799112
Babu MM, van der Lee R, de Groot NS, Gsponer J (2011) Intrinsically disordered proteins: regulation and disease. Curr Opin Struct Biol 21(3):432–440. https://doi.org/10.1016/j.sbi.2011.03.011
Babu MM (2016) The contribution of intrinsically disordered regions to protein function, cellular complexity, and human disease. Biochem Soc Trans 44(5):1185–1200. https://doi.org/10.1042/BST20160172
Baggio F, Bozzato A, Benna C, Leonardi E, Romoli O, Cognolato M, Tosatto SC, Costa R, Sandrelli F (2013) 2mit, an intronic gene of Drosophila melanogaster timeless2, is involved in behavioral plasticity. PLoS One 8(9):e76351. https://doi.org/10.1371/journal.pone.0076351
Barabasi AL, Albert R (1999) Emergence of scaling in random networks. Science 286(5439):509–512. https://doi.org/10.1126/science.286.5439.509
Barabási AL (2009) Scale-free networks: a decade and beyond. Science 325(5939):412–413. https://doi.org/10.1126/science.1173299
Boehr DD, Nussinov R, Wright PE (2009) The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5(11):789–796. https://doi.org/10.1038/nchembio.232
Borgia A, Borgia MB, Bugge K, Kissling VM, Heidarsson PO, Fernandes CB, Sottini A, Soranno A, Buholzer KJ, Nettels D, Kragelund BB, Best RB, Schuler B (2018) Extreme disorder in an ultrahigh-affinity protein complex. Nature 555(7694):61–66. https://doi.org/10.1038/nature25762
Brock A, Chang H, Huang S (2009) Non-genetic heterogeneity–a mutation-independent driving force for the somatic evolution of tumours. Nat Rev Genet 10(5):336–342. https://doi.org/10.1038/nrg2556
Bürgi J, Xue B, Uversky VN, van der Goot FG (2016) Intrinsic disorder in transmembrane proteins: roles in signaling and topology prediction. PLoS One 11(7):e0158594. https://doi.org/10.1371/journal.pone.0158594
Chakrabortee S, Meersman F, Kaminski Schierle GS, Bertoncini CW, McGee B, Kaminski CF, Tunnacliffe A (2010) Catalytic and chaperone-like functions in an intrinsically disordered protein associated with desiccation tolerance. Proc Natl Acad Sci U S A 107(37):16084–16089. https://doi.org/10.1073/pnas.1006276107
Choi UB, McCann JJ, Weninger KR, Bowen ME (2011) Beyond the random coil: stochastic conformational switching in intrinsically disordered proteins. Structure 19(4):566–576. https://doi.org/10.1016/j.str.2011.01.011
Choi UB, Sanabria H, Smirnova T, Bowen ME, Weninger KR (2019) Spontaneous switching among conformational ensembles in intrinsically disordered proteins. Biomolecules 9(3):114. https://doi.org/10.3390/biom9030114
Chung KM, Kolling FW 4th, Gajdosik MD, Burger S, Russell AC, Nelson CE (2014) Single cell analysis reveals the stochastic phase of reprogramming to pluripotency is an ordered probabilistic process. PLoS One. 9(4):e95304. https://doi.org/10.1371/journal.pone.0095304
Dehmamy N, Milanlouei S, Barabási AL (2018) A structural transition in physical networks. Nature 563(7733):676–680. https://doi.org/10.1038/s41586-018-0726-6
Dong P, Fan Y, Sun J, Lv M, Yi M, Tan X, Liu S (2016) A dynamic interaction process between KaiA and KaiC is critical to the cyanobacterial circadianoscillator. Sci Rep 6:25129. https://doi.org/10.1038/srep25129
Dosztányi Z, Chen J, Dunker AK, Simon I, Tompa P (2006) Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res 5(11):2985–2995. https://doi.org/10.1021/pr060171o
Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z (2001) Intrinsically disordered protein. J Mol Graph Model 19(1):26–59. https://doi.org/10.1016/s1093-3263(00)00138-8
Dyson HJ, Wright PE (2002) Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol 12(1):54–60. https://doi.org/10.1016/s0959-440x(02)00289-0
Dyson HJ, Wright PE (2021) NMR illuminates intrinsic disorder. Curr Opin Struct Biol 2(70):44–52. https://doi.org/10.1016/j.sbi.2021.03.015
Edwards YJ, Lobley AE, Pentony MM, Jones DT (2009) Insights into the regulation of intrinsically disordered proteins in the human proteome by analyzing sequence and gene expression data. Genome Biol 10(5):R50. https://doi.org/10.1186/gb-2009-10-5-r50
Eldar A, Elowitz MB (2010) Functional roles for noise in genetic circuits. Nature 467(7312):167–173. https://doi.org/10.1038/nature09326
Fischer KR, Durrans A, Lee S, Sheng J, Li F, Wong ST, Choi H, El Rayes T, Ryu S, Troeger J, Schwabe RF, Vahdat LT, Altorki NK, Mittal V, Gao D (2015) Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 527(7579):472–476. https://doi.org/10.1038/nature15748
Freiberger MI, Wolynes PG, Ferreiro DU, Fuxreiter M (2021) Frustration in fuzzy protein complexes leads to interaction versatility. J Phys Chem B 125(10):2513–2520. https://doi.org/10.1021/acs.jpcb.0c11068
Fuxreiter M (2018) Towards a stochastic paradigm: from fuzzy ensembles to cellular functions. Molecules 23(11):pii E3008. https://doi.org/10.3390/molecules23113008
Fuxreiter M (2020) Fuzzy protein theory for disordered proteins. Biochem Soc Trans 48(6):2557–2564. https://doi.org/10.1042/BST20200239
Galea CA, Wang Y, Sivakolundu SG, Kriwacki RW (2008) Regulation of cell division by intrinsically unstructured proteins: intrinsic flexibility, modularity, and signaling conduits. Biochemistry 47(29):7598–7609. https://doi.org/10.1021/bi8006803
George JT, Jolly MK, Xu S, Somarelli JA, Levine H (2017) Survival outcomes in cancer patients predicted by a partial EMT gene expression scoring metric. Cancer Res 77(22):6415–6428. https://doi.org/10.1158/0008-5472.CAN-16-3521
Gsponer J, Babu MM (2009) The rules of disorder or why disorder rules. Prog Biophys Mol Biol 99(2–3):94–103. https://doi.org/10.1016/j.pbiomolbio.2009.03.001
Gsponer J, Futschik ME, Teichmann SA, Babu MM (2008) Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science 322(5906):1365–1368. https://doi.org/10.1126/science.1163581
Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C, Lander ES (2011) Stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell 146(4):633–644. https://doi.org/10.1016/j.cell.2011.07.026
Hammerlindl H, Schaider H (2018) Tumor cell-intrinsic phenotypic plasticity facilitates adaptive cellular reprogramming driving acquired drug resistance. J Cell Commun Signal 12(1):133–141. https://doi.org/10.1007/s12079-017-0435-1
Hansen MMK, Desai RV, Simpson ML, Weinberger LS (2018) Cytoplasmic amplification of transcriptional noise generates substantial cell-to-cell variability. Cell Syst 7(4):384–397. https://doi.org/10.1016/j.cels.2018.08.002
Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM (2006) Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol 2(8):e100. https://doi.org/10.1371/journal.pcbi.0020100
He Y, Chen Y, Mooney SM, Rajagopalan K, Bhargava A, Sacho E, Weninger K, Bryan PN, Kulkarni P, Orban J (2015) Phosphorylation-induced conformational ensemble switching in an intrinsically disordered cancer/testis antigen. J Biol Chem 290(41):25090–25102. https://doi.org/10.1074/jbc.M115.658583
Hu G, Wu Z, Uversky VN, Kurgan L (2017) Functional analysis of human hub proteins and their interactors involved in the intrinsic disorder-enriched interactions. Int J Mol Sci 18(12):pii: E2761. https://doi.org/10.3390/ijms18122761
Huang S (2009) Non-genetic heterogeneity of cells in development: more than just noise. Development 136:3853–3862. https://doi.org/10.1242/dev.035139
Hurley JM, Larrondo LF, Loros JJ, Dunlap JC (2013) Conserved RNA helicase FRH acts nonenzymatically to support the intrinsically disordered neurospora clock protein FRQ. Mol Cell 52(6):832–843. https://doi.org/10.1016/j.molcel.2013.11.005
Hurley JM, Loros JJ, Dunlap JC (2016) Circadian oscillators: around the transcription-translation feedback loop and on to output. Trends Biochem Sci 41(10):834–846. https://doi.org/10.1016/j.tibs.2016.07.009
Iakoucheva LM, Brown CJ, Lawson JD, Obradović Z, Dunker AK (2002) Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol 323(3):573–584. https://doi.org/10.1016/s0022-2836(02)00969-5
Jia D, Jolly MK, Kulkarni P, Levine H (2017) Phenotypic plasticity and cell fate decisions in cancer: insights from dynamical systems theory. Cancers (basel) 9(7):70. https://doi.org/10.3390/cancers9070070
Karacosta LG, Anchang B, Ignatiadis N, Kimmey SC, Benson JA, Shrager JB, Tibshirani R, Bendall SC, Plevritis SK (2019) Mapping lung cancer epithelial-mesenchymal transition states and trajectories with single-cell resolution. Nat Commun 10(1):5587. https://doi.org/10.1038/s41467-019-13441-6
Kontogeorgaki S, Sánchez-García RJ, Ewing RM, Zygalakis KC, MacArthur BD (2017) Noise-Processing by Signaling Networks Sci Rep 7(1):532. https://doi.org/10.1038/s41598-017-00659-x
Kulkarni P, Jolly MK, Jia D, Mooney SM, Bhargava A, Kagohara LT, Chen Y, Hao P, He Y, Veltri RW, Grishaev A, Weninger K, Levine H, Orban J (2017) Phosphorylation-induced conformational dynamics in an intrinsically disordered protein and potential role in phenotypic heterogeneity. Proc Natl Acad Sci U S A 114(13):E2644–E2653. https://doi.org/10.1073/pnas.1700082114
Kulkarni V, Kulkarni P (2019) Intrinsically disordered proteins and phenotypic switching: implications in cancer. Prog Mol Biol Transl Sci 166:63–84. https://doi.org/10.1016/bs.pmbts.2019.03.013
Kumar N, Cramer GM, Dahaj SAZ, Sundaram B, Celli JP, Kulkarni RV (2019) Stochastic modeling of phenotypic switching and chemoresistance in cancer cell populations. Sci Rep 9(1):10845. https://doi.org/10.1038/s41598-019-46926-x
Kuwahara H, Gao X (2013) Stochastic effects as a force to increase the complexity of signaling networks. Sci Rep 3:2297. https://doi.org/10.1038/srep02297
Ladbury JE, Arold ST (2012) Noise in cellular signaling pathways: causes and effects. Trends Biochem Sci 37(5):173–178. https://doi.org/10.1016/j.tibs.2012.01.001
Lin X, Kulkarni P, Bocci F, Schafer NP, Roy S, Tsai MY, He Y, Chen Y, Rajagopalan K, Mooney SM, Zeng Y, Weninger K, Grishaev A, Onuchic JN, Levine H, Wolynes PG, Salgia R, Rangarajan G, Uversky V, Orban J, Jolly MK (2019) Structural and dynamical order of a disordered protein: molecular insights into conformational switching of PAGE4 at the systems level. Biomolecules 9(2):77. https://doi.org/10.3390/biom9020077
Lin YT, Hufton PG, Lee EJ, Potoyan DA (2018) A stochastic and dynamical view of pluripotency in mouse embryonic stem cells. PLoS Comput Biol 14(2):e1006000. https://doi.org/10.1371/journal.pcbi.1006000
Lv C, Fu S, Dong Q, Yu Z, Zhang G, Kong C, Fu C, Zeng Y (2019) PAGE4 promotes prostate cancer cells survive under oxidative stress through modulating MAPK/JNK/ERK pathway. J Exp Clin Cancer Res 38(1):24. https://doi.org/10.1186/s13046-019-1032-3
MacArthur BD, Please CP, Oreffo RO (2008) Stochasticity and the molecular mechanisms of induced pluripotency. PLoS One 3(8):e3086. https://doi.org/10.1371/journal.pone.0003086
Mahmoudabadi G, Rajagopalan K, Getzenberg RH, Hannenhalli S, Rangarajan G, Kulkarni P (2013) Intrinsically disordered proteins and conformational noise: implications in cancer. Cell Cycle 12(1):26–31. https://doi.org/10.4161/cc.23178
Marcotte EM, Tsechansky M (2009) Disorder, promiscuity, and toxic partnerships. Cell 138(1):16–18. https://doi.org/10.1016/j.cell.2009.06.024
Michael AK, Fribourgh JL, Van Gelder RN, Partch CL (2017) Animal cryptochromes: divergent roles in light perception, circadian timekeeping and beyond. Photochem Photobiol 93(1):128–140. https://doi.org/10.1111/php.12677
Mitrea DM, Yoon MK, Ou L, Kriwacki RW (2012) Disorder-function relationships for the cell cycle regulatory proteins p21 and p27. Biol Chem 393(4):259–274. https://doi.org/10.1515/hsz-2011-0254
Mooney SM, Jolly MK, Levine H, Kulkarni P (2016) Phenotypic plasticity in prostate cancer: role of intrinsically disordered proteins. Asian J Androl 18(5):704–710. https://doi.org/10.4103/1008-682X.183570
Mooney SM, Qiu R, Kim JJ, Sacho EJ, Rajagopalan K, Johng D, Shiraishi T, Kulkarni P, Weninger KR (2014) Cancer/testis antigen PAGE4, a regulator of c-Jun transactivation, is phosphorylated by homeodomain-interacting protein kinase 1, a component of the stress-response pathway. Biochemistry 53(10):1670–1679. https://doi.org/10.1021/bi500013w
Mylona A, Theillet FX, Foster C, Cheng TM, Miralles F, Bates PA, Selenko P, Treisman R (2016) Opposing effects of Elk-1 multisite phosphorylation shape its response to ERK activation. Science 354(6309):233–237. https://doi.org/10.1126/science.aad1872
Nichol D, Robertson-Tessi M, Jeavons P, Anderson AR (2016) Stochasticity in the genotype-phenotype map: implications for the robustness and persistence of bet-hedging. Genetics 204(4):1523–1539. https://doi.org/10.1534/genetics.116.193474
Patil A, Kinoshita K, Nakamura H (2010) Hub promiscuity in protein-protein interaction networks. Int J Mol Sci 11(4):1930–1943. https://doi.org/10.3390/ijms11041930
Peng Z, Yan J, Fan X, Mizianty MJ, Xue B, Wang K, Hu G, Uversky VN, Kurgan L (2015) Exceptionally abundant exceptions: comprehensive characterization of intrinsic disorder in all domains of life. Cell Mol Life Sci 72(1):137–151. https://doi.org/10.1007/s00018-014-1661-9
Rambow F, Marine JC, Goding CR (2019) Melanoma plasticity and phenotypic diversity: therapeutic barriers and opportunities. Genes Dev 33(19–20):1295–1318. https://doi.org/10.1101/gad.329771.119
Rangarajan N, Fox Z, Singh A, Kulkarni P, Rangarajan G (2015) Disorder, oscillatory dynamics and state switching: the role of c-Myc. J Theor Biol 386:105–114. https://doi.org/10.1016/j.jtbi.2015.09.013
Rangarajan N, Kulkarni P, Hannenhalli S (2015) Evolutionarily conserved network properties of intrinsically disordered proteins. PLoS One 10(5):e0126729. https://doi.org/10.1371/journal.pone.0126729
Raj A, van Oudenaarden A (2009) Single-molecule approaches to stochastic gene expression. Annu Rev Biophys 38:255–270. https://doi.org/10.1146/annurev.biophys.37.032807.125928
Rajagopalan K, Mooney SM, Parekh N, Getzenberg RH, Kulkarni P (2011) A majority of the cancer/testis antigens are intrinsically disordered proteins. J Cell Biochem 112(11):3256–3267. https://doi.org/10.1002/jcb.23252
Rajagopalan K, Qiu R, Mooney SM, Rao S, Shiraishi T, Sacho E, Huang H, Shapiro E, Weninger KR, Kulkarni P (2014) The stress-response protein prostate-associated gene 4, interacts with c-Jun and potentiates its transactivation. Biochim Biophys Acta 1842(2):154–163. https://doi.org/10.1016/j.bbadis.2013.11.014
Ruscetti M, Dadashian EL, Guo W, Quach B, Mulholland DJ, Park JW, Tran LM, Kobayashi N, Bianchi-Frias D, Xing Y, Nelson PS, Wu H (2016) HDAC inhibition impedes epithelial-mesenchymal plasticity and suppresses metastatic, castration-resistant prostate cancer. Oncogene 35(29):3781–3795. https://doi.org/10.1038/onc.2015.444
Safdari H, Kalirad A, Picioreanu C, Tusserkani R, Goliaei B, Sadeghi M (2020) Noise-driven cell differentiation and the emergence of spatiotemporal patterns. PLoS One. 15(4):e0232060. https://doi.org/10.1371/journal.pone.0232060
Sahoo S, Mishra A, Kaur H, Hari K, Muralidharan S, Mandal S, Jolly MK (2021) A mechanistic model captures the emergence and implications of non-genetic heterogeneity and reversible drug resistance in ER+ breast cancer cells. NAR Cancer 3(3):zcab027. https://doi.org/10.1093/narcan/zcab027
Sato N, Sadar MD, Bruchovsky N, Saatcioglu F, Rennie PS, Sato S, Lange PH, Gleave ME (1997) Androgenic induction of prostate-specific antigen gene is repressed by protein-protein interaction between the androgen receptor and AP-1/c-Jun in the human prostate cancer cell line LNCaP. J Biol Chem 272(28):17485–17494. https://doi.org/10.1074/jbc.272.28.17485
Salgia R, Jolly MK, Dorff T, Lau C, Weninger K, Orban J, Kulkarni P (2018) Prostate-associated gene 4 (PAGE4): leveraging the conformational dynamics of a dancing protein cloud as a therapeutic target. J Clin Med 7(6):156. https://doi.org/10.3390/jcm7060156
Shachaf CM, Kopelman AM, Arvanitis C, Karlsson A, Beer S, Mandl S, Bachmann MH, Borowsky AD, Ruebner B, Cardiff RD, Yang Q, Bishop JM, Contag CH, Felsher DW (2004) MYC inactivation uncovers pluripotent differentiation and tumour dormancy in hepatocellular cancer. Nature 431(7012):1112–1117. https://doi.org/10.1038/nature03043
Shachaf CM, Felsher DW (2005) Tumor dormancy and MYC inactivation: pushing cancer to the brink of normalcy. Cancer Res 65(11):4471–4474. https://doi.org/10.1158/0008-5472.CAN-05-1172
Shammas SL (2017) Mechanistic roles of protein disorder within transcription. Curr Opin Struct Biol 42:155–161. https://doi.org/10.1016/j.sbi.2017.02.003
Sehl ME, Shimada M, Landeros A, Lange K, Wicha MS (2015) Modeling of cancer stem cell state transitions predicts therapeutic response. PLoS One 10(9):e0135797. https://doi.org/10.1371/journal.pone.0135797
Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, Wong KK, Brandstetter K, Wittner B, Ramaswamy S, Classon M, Settleman J (2010) A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141(1):69–80. https://doi.org/10.1016/j.cell.2010.02.027
Sharma R, Raduly Z, Miskei M, Fuxreiter M (2015) Fuzzy Complexes: specific binding without complete folding. FEBS Lett 589(19 Pt A):2533–42. https://doi.org/10.1016/j.febslet.2015.07.022
Singh D, Bocci F, Kulkarni P, Jolly MK (2021) Coupled feedback loops involving PAGE4, EMT and Notch signaling can give rise to non-genetic heterogeneity in prostate cancer cells. Entropy (basel) 23(3):288. https://doi.org/10.3390/e23030288
Smith Q, Stukalin E, Kusuma S, Gerecht S, Sun SX (2015) Stochasticity and spatial interaction govern stem cell differentiation dynamics. Sci Rep 31(5):12617. https://doi.org/10.1038/srep12617
Sonnenschein C, Soto AM, Rangarajan A, Kulkarni P (2014) Competing views on cancer. J Biosci 39(2):281–302. https://doi.org/10.1007/s12038-013-9403-y
Su Y, Wei W, Robert L, Xue M, Tsoi J, Garcia-Diaz A, Homet Moreno B, Kim J, Ng RH, Lee JW, Koya RC, Comin-Anduix B, Graeber TG, Ribas A, Heath JR (2017) Single-cell analysis resolves the cell state transition and signaling dynamics associated with melanoma drug-induced resistance. Proc Natl Acad Sci U S A 114(52):13679–13684. https://doi.org/10.1073/pnas.1712064115
Sugase K, Dyson HJ, Wright PE (2007) Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature 447(7147):1021–1025. https://doi.org/10.1038/nature05858
Tillman K, Oberfield JL, Shen XQ, Bubulya A, Shemshedini L (1998) c-Fos dimerization with c-Jun represses c-Jun enhancement of androgen receptor transactivation. Endocrine 9(2):193–200. https://doi.org/10.1385/ENDO:9:2:193
Tompa P (2011) Unstructural biology coming of age. Curr Opin Struct Biol 21(3):419–425. https://doi.org/10.1016/j.sbi.2011.03.012
Uversky VN, Oldfield CJ, Dunker AK (2008) Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys 37:215–246. https://doi.org/10.1146/annurev.biophys.37.032807.125924
Uversky VN, Dunker AK (2010) Understanding protein non-folding. Biochim Biophys Acta 1804(6):1231–1264. https://doi.org/10.1016/j.bbapap.2010.01.017
Uversky VN (2014) Wrecked regulation of intrinsically disordered proteins in diseases: pathogenicity of deregulated regulators. Front Mol Biosci 1:6. https://doi.org/10.3389/fmolb.2014.00006
Uversky VN (2015) The multifaceted roles of intrinsic disorder in protein complexes. FEBS Lett 589(19 Pt A):2498–506. https://doi.org/10.1016/j.febslet.2015.06.004
Uversky VN (2017) Paradoxes and wonders of intrinsic disorder: stability of instability. Intrinsically Disord Proteins. 5(1):e1327757. https://doi.org/10.1080/21690707.2017.1327757
Uversky VN (2018) Intrinsic disorder, protein-protein interactions, and disease. Adv Protein Chem Struct Biol 110:85–121. https://doi.org/10.1016/bs.apcsb.2017.06.005
Uversky VN (2021) Per aspera ad chaos: a personal journey to the wonderland of intrinsic disorder. Biochem J 478(15):3015–3030. https://doi.org/10.1042/BCJ20210146
Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B (2009) Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138(1):198–208. https://doi.org/10.1016/j.cell.2009.04.029
Wakao S, Kitada M, Dezawa M (2013) The elite and stochastic model for iPS cell generation: multilineage-differentiating stress enduring (Muse) cells are readily reprogrammable into iPS cells. Cytometry A 83(1):18–26. https://doi.org/10.1002/cyto.a.22069
Wang Y, Chu X, Longhi S, Roche P, Han W, Wang E, Wang J (2013) Multiscaled exploration of coupled folding and binding of an intrinsically disordered molecular recognition element in measles virus nucleoprotein. Proc Natl Acad Sci U S A 110(40):E3743–E3752. https://doi.org/10.1073/pnas.1308381110
Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT (2004) Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol 337(3):635–645. https://doi.org/10.1016/j.jmb.2004.02.002
Wright PE, Dyson HJ (1999) Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol 293(2):321–331. https://doi.org/10.1006/jmbi.1999.3110
Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16(1):18–29. https://doi.org/10.1038/nrm3920
Xue B, Oldfield CJ, Van YY, Dunker AK, Uversky VN (2012) Protein intrinsic disorder and induced pluripotent stem cells. Mol Biosyst 8(1):134–150. https://doi.org/10.1039/c1mb05163f
Xue B, Dunker AK, Uversky VN (2012) Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J Biomol Struct Dyn 30(2):137–149. https://doi.org/10.1080/07391102.2012.675145
Yamanaka S (2009) Elite and stochastic models for induced pluripotent stem cell generation. Nature 460:49–52. https://doi.org/10.1038/nature08180
Yoon MK, Mitrea DM, Ou L, Kriwacki RW (2012) Cell cycle regulation by the intrinsically disordered proteins p21 and p27. Biochem Soc Trans 40(5):981–988. https://doi.org/10.1042/BST20120092
Zeng Y, He Y, Yang F, Mooney SM, Getzenberg RH, Orban J, Kulkarni P (2011) The cancer/testis antigen prostate-associated gene 4 (PAGE4) is a highly intrinsically disordered protein. J Biol Chem 286(16):13985–13994. https://doi.org/10.1074/jbc.M110.210765
Zeng Y, Gao D, Kim JJ, Shiraishi T, Terada N, Kakehi Y, Kong C, Getzenberg RH, Kulkarni P (2013) Prostate-associated gene 4 (PAGE4) protects cells against stress by elevating p21 and suppressing reactive oxygen species production. Am J Clin Exp Urol 1(1):39–52
Zheng X, Carstens JL, Kim J, Scheible M, Kaye J, Sugimoto H, Wu CC, LeBleu VS, Kalluri R (2015) Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 527(7579):525–530. https://doi.org/10.1038/nature16064
Acknowledgements
PK thanks Dr. Amita Behal for her thoughtful comments on a previous version of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest/Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kulkarni, P., Achuthan, S., Bhattacharya, S. et al. Protein conformational dynamics and phenotypic switching. Biophys Rev 13, 1127–1138 (2021). https://doi.org/10.1007/s12551-021-00858-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12551-021-00858-x