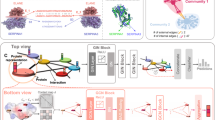
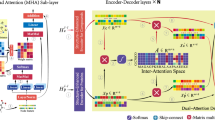
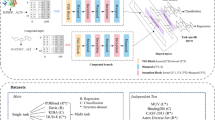
Abstract
Gathering information from multi-perspective graphs is an essential issue for many applications especially for protein–ligand-binding affinity prediction. Most of traditional approaches obtained such information individually with low interpretability. In this paper, we harness the rich information from multi-perspective graphs with a general model, which abstractly represents protein–ligand complexes with better interpretability while achieving excellent predictive performance. In addition, we specially analyze the protein–ligand-binding affinity problem, taking into account the heterogeneity of proteins and ligands. Experimental evaluations demonstrate the effectiveness of our data representation strategy on public datasets by fusing information from different perspectives. All codes are available in the https://github.com/Jthy-af/HaPPy.
Graphical Abstract









Similar content being viewed by others
Data availability
Data openly available in a public repository. The data that support the findings of this study are openly available in PDBbind dataset at http://pdbbind.org.cn. And all codes are available in the https://github.com/Jthy-af/HaPPy or from the corresponding author on reasonable request.
References
Kitchen DB, Decornez H, Furr JR et al (2004) Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discovery 3(11):935–949. https://doi.org/10.1038/nrd1549
Du BX, Qin Y, Jiang YF et al (2022) Compound-protein interaction prediction by deep learning: databases, descriptors and models. Drug Discovery Today 27(5):1350–1366. https://doi.org/10.1016/j.drudis.2022.02.023
Ferreira LG, Dos Santos RN, Oliva G et al (2015) Molecular docking and structure-based drug design strategies. Molecules 20(7):13384–13421. https://doi.org/10.3390/molecules200713384
Pinzi L, Rastelli G (2019) Molecular docking: shifting paradigms in drug discovery. Int J Mol Sci 20(18):4331. https://doi.org/10.3390/ijms20184331
Sethi A, Joshi K, Sasikala K et al (2019) Molecular docking in modern drug discovery: principles and recent applications. In: Drug Discovery and Development - New Advances, vol 2, pp. 1–21. https://doi.org/10.5772/intechopen.85991
Li J, Fu A, Zhang L (2019) An overview of scoring functions used for protein-ligand interactions in molecular docking. Interdiscip Sci Comput Life Sci 11(2):320–328. https://doi.org/10.1007/s12539-019-00327-w
Khamis MA, Gomaa W, Ahmed WF (2015) Machine learning in computational docking. Artif Intell Med 63(3):135–152. https://doi.org/10.1016/j.artmed.2015.02.002
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 770–778. https://doi.org/10.1109/CVPR.2016.90
Devlin J, Chang MW, Lee K et al (2019) Bert: Pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), pp. 4171–4186. https://doi.org/10.18653/v1/N19-1423
Chang CH, Hung CL, Tang CY (2019) A review of deep learning in computer-aided drug design. In: 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 1856–1861. https://doi.org/10.1109/BIBM47256.2019.8982968
Xie T, Grossman JC (2018) Crystal graph convolutional neural networks for an accurate and interpretable prediction of material properties. Phys Rev Lett 120(14):145301. https://doi.org/10.1103/PhysRevLett.120.145301
Ragoza M, Hochuli J, Idrobo E et al (2017) Protein-ligand scoring with convolutional neural networks. J Chem Inf Model 57(4):942–957. https://doi.org/10.1021/acs.jcim.6b00740
Stepniewska-Dziubinska MM, Zielenkiewicz P, Siedlecki P (2018) Development and evaluation of a deep learning model for protein-ligand binding affinity prediction. Bioinformatics 34(21):3666–3674. https://doi.org/10.1093/bioinformatics/bty374
Rezaei MA, Li Y, Wu D et al (2020) Deep learning in drug design: protein-ligand binding affinity prediction. IEEE/ACM Trans Comput Biol Bioinf 19(1):407–417. https://doi.org/10.1109/TCBB.2020.3046945
Öztürk H, Özgür A, Ozkirimli E (2018) Deepdta: deep drug-target binding affinity prediction. Bioinformatics 34(17):821–829. https://doi.org/10.1093/bioinformatics/bty593
Wang K, Zhou R, Li Y et al (2021) Deepdtaf: a deep learning method to predict protein-ligand binding affinity. Brief Bioinform 22(5):072. https://doi.org/10.1093/bib/bbab072
Wang W, Sun B, Liu D et al (2021) Dpla: prediction of protein-ligand binding affinity by integrating multi-level information. In: 2021 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), pp. 3428–3434. https://doi.org/10.1109/BIBM52615.2021.9669576
Lim J, Ryu S, Park K et al (2019) Predicting drug-target interaction using a novel graph neural network with 3d structure-embedded graph representation. J Chem Inf Model 59(9):3981–3988. https://doi.org/10.1021/acs.jcim.9b00387
Li S, Zhou J, Xu T et al (2021) Structure-aware interactive graph neural networks for the prediction of protein-ligand binding affinity. In: Proceedings of the 27th ACM SIGKDD Conference on Knowledge Discovery & Data Mining, pp. 975–985. https://doi.org/10.1145/3447548.3467311
Jiang D, Hsieh C-Y, Wu Z et al (2021) Interactiongraphnet: a novel and efficient deep graph representation learning framework for accurate protein-ligand interaction predictions. J Med Chem 64(24):18209–18232. https://doi.org/10.1021/acs.jmedchem.1c01830
Nguyen T, Le H, Quinn TP et al (2021) Graphdta: predicting drug-target binding affinity with graph neural networks. Bioinformatics 37(8):1140–1147. https://doi.org/10.1093/bioinformatics/btaa921
Zheng L, Fan J, Mu Y (2019) Onionnet: a multiple-layer intermolecular-contact-based convolutional neural network for protein-ligand binding affinity prediction. ACS Omega 4(14):15956–15965. https://doi.org/10.1021/acsomega.9b01997
Su M, Yang Q, Du Y et al (2019) Comparative assessment of scoring functions: the casf-2016 update. J Chem Inf Model 59(2):895–913. https://doi.org/10.1021/acs.jcim.8b00545
Klicpera J, Groß J, Günnemann S (2020) Directional message passing for molecular graphs. arXiv preprint arXiv:2003.03123https://doi.org/10.48550/arXiv.2003.03123
Trott O, Olson AJ (2010) Autodock vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31(2):455–461. https://doi.org/10.1002/jcc.21334
Bajusz D, Ferenczy GG, Keseru GM (2017) Structure-based virtual screening approaches in kinase-directed drug discovery. Curr Top Med Chem 17(20):2235–2259. https://doi.org/10.2174/1568026617666170224121313
Ballester PJ, Mitchell JB (2010) A machine learning approach to predicting protein-ligand binding affinity with applications to molecular docking. Bioinformatics 26(9):1169–1175. https://doi.org/10.1093/bioinformatics/btq112
Kinnings SL, Liu N, Tonge PJ et al (2011) A machine learning-based method to improve docking scoring functions and its application to drug repurposing. J Chem Inf Model 51(2):408–419. https://doi.org/10.1021/ci100369f
Wee J, Xia K (2021) Ollivier persistent ricci curvature-based machine learning for the protein-ligand binding affinity prediction. J Chem Inf Model 61(4):1617–1626. https://doi.org/10.1021/acs.jcim.0c01415
Durrant JD, McCammon JA (2010) Nnscore: a neural-network-based scoring function for the characterization of protein- ligand complexes. J Chem Inf Model 50(10):1865–1871. https://doi.org/10.1021/ci100244v
Weininger D (1988) Smiles a chemical language and information system 1 introduction to methodology and encoding rules. J Chem Inf Comput Sci 28(1):31–36. https://doi.org/10.1021/ci00057a005
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv preprint arXiv:1609.02907https://doi.org/10.48550/arXiv.1609.02907
Veličcković P, Cucurull G, Casanova A et al (2017) Graph attention networks. arXiv preprint arXiv:1710.10903, https://doi.org/10.48550/arXiv.1710.10903
Wang X, Ji H, Shi C et al (2019) Heterogeneous graph attention network. In: The World Wide Web Conference, pp. 2022–2032 https://doi.org/10.1145/3308558.3313562
Xie T, He C, Ren X et al (2022) L-bgnn: layerwise trained bipartite graph neural networks. IEEE Transact Neural Netw Learn Syst 1–13. https://doi.org/10.1109/TNNLS.2022.3171199
Xu K, Hu W, Leskovec J et al (2018) How powerful are graph neural networks? arXiv preprint arXiv:1810.00826, https://doi.org/10.48550/arXiv.1810.00826
Xiong Z, Wang D, Liu X et al (2019) Pushing the boundaries of molecular representation for drug discovery with the graph attention mechanism. J Med Chem 63(16):8749–8760. https://doi.org/10.1021/acs.jmedchem.9b00959
Elnaggar A, Heinzinger M, Dallago C et al (2020) Prottrans: towards cracking the language of life’s code through self-supervised deep learning and high performance computing. arXiv preprint arXiv:2007.06225, https://doi.org/10.48550/arXiv.2007.06225
Wang R, Fang X, Lu Y et al (2005) The pdbbind database: methodologies and updates. J Med Chem 48(12):4111–4119. https://doi.org/10.1021/jm048957q
Jin W, Barzilay R, Jaakkola T (2018) Junction tree variational autoencoder for molecular graph generation. In: International Conference on Machine Learning, pp. 2323–2332. https://doi.org/10.1039/9781788016841-00228
Acknowledgements
This work is supported by National Natural Science Foundation of China under Grant 22033002 and Grant 92370127, in part by National Natural Science Foundation of China under Grant 21873050.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Li, Y., Wang, J. et al. A Multi-perspective Model for Protein–Ligand-Binding Affinity Prediction. Interdiscip Sci Comput Life Sci 15, 696–709 (2023). https://doi.org/10.1007/s12539-023-00582-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-023-00582-y