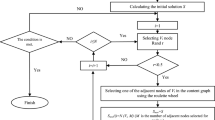
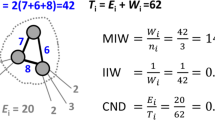
Abstract
The widespread availability and importance of large-scale protein-protein interaction (PPI) data demand a flurry of research efforts to understand the organisation of a cell and its functionality by analysing these data at the network level. In the bioinformatics and data mining fields, network clustering acquired a lot of attraction to examine a PPI network’s topological and functional aspects. The clustering of PPI networks has been proven to be an excellent method for discovering functional modules, disclosing functions of unknown proteins, and other tasks in numerous research over the last decade. This research proposes a unique graph mining approach to detect protein complexes using dense neighbourhoods (highly connected regions) in an interaction graph. Our technique first finds size-3 cliques associated with each edge (protein interaction), and then these core cliques are expanded to form high-density subgraphs. Loosely connected proteins are stripped out from these subgraphs to produce a potential protein complex. Finally, the redundancy is removed based on the Jaccard coefficient. Computational results are presented on the yeast and human protein interaction dataset to highlight our proposed technique’s efficiency. Predicted protein complexes of the proposed approach have a significantly higher score of similarity to those used as gold standards in the CYC-2008 and CORUM benchmark databases than other existing approaches.
Graphical Abstract





Similar content being viewed by others
References
Patra S, Mohapatra A (2020) Review of tools and algorithms for network motif discovery in biological networks. IET Syst Biol 14(4):171–189. https://doi.org/10.1049/iet-syb.2020.0004
Grigorov MG (2005) Global properties of biological networks. Drug Discov Today 10(5):365–72. https://doi.org/10.1016/S1359-6446(05)03369-6
Khanin R, Wit E (2006) How scale-free are biological networks. J Comput Biol 13(3):810–818. https://doi.org/10.1089/cmb.2006.13.810
Barabasi AL, Albert R (1999) Emergence of scaling in random networks. Science 286(5439):509–512. https://doi.org/10.1126/science.286.5439.509
Balasundaram B, Butenko S, Trukhanov S (2005) Novel approaches for analyzing biological networks. J Comb Optim 10(1):23–39. https://doi.org/10.1007/s10878-005-1857-x
Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868):141–147. https://doi.org/10.1038/415141a
Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440(7084):631–636. https://doi.org/10.1038/nature04532
Song L, Li D, Zeng X, Wu Y, Guo L, Zou Q (2014) nDNA-prot: identification of DNA-binding proteins based on unbalanced classification. BMC Bioinform 15(1):1–10. https://doi.org/10.1186/1471-2105-15-298
Cao B, Luo J, Liang C, Wang S, Song D (2015) Moepga: A novel method to detect protein complexes in yeast protein-protein interaction networks based on multiobjective evolutionary programming genetic algorithm. Comput Biol Chem 58:173–181. https://doi.org/10.1016/j.compbiolchem.2015.06.006
ur Rehman Z, Idris A, Khan A (2018) Multi-dimensional scaling based grouping of known complexes and intelligent protein complex detection. Comput Biol Chem 74:149–156. https://doi.org/10.1016/j.compbiolchem.2018.03.023
Zahiri J, Emamjomeh A, Bagheri S, Ivazeh A, Mahdevar G, Tehrani HS, Mirzaie M, Fakheri BA, Mohammad-Noori M (2020) Protein complex prediction: a survey. Genomics 112(1):174–183. https://doi.org/10.1016/j.ygeno.2019.01.011
Zaslavsky L, Ciufo S, Fedorov B, Tatusova T (2016) Clustering analysis of proteins from microbial genomes at multiple levels of resolution. BMC Bioinform 17(8):545–552. https://doi.org/10.1186/s12859-016-1112-8
Yu L, Gao L, Li K, Zhao Y, Chiu DK (2011) A degree-distribution based hierarchical agglomerative clustering algorithm for protein complexes identification. Comput Biol Chem 35(5):298–307. https://doi.org/10.1016/j.compbiolchem.2011.07.005
Hartwell LH, Hopfield JJ, Leibler S, Murray AW (1999) From molecular to modular cell biology. Nature 402(6761):C47–C52. https://doi.org/10.1038/35011540
Keretsu S, Sarmah R (2016) Weighted edge based clustering to identify protein complexes in protein-protein interaction networks incorporating gene expression profile. Comput Biol Chem 65:69–79. https://doi.org/10.1016/j.compbiolchem.2016.10.001
Jalili S, Marashi SA (2015) CAMWI: detecting protein complexes using weighted clustering coefficient and weighted density. Comput Biol Chem 58:231–240. https://doi.org/10.1016/j.compbiolchem.2015.07.012
Pyrogova I, Wong L (2018) Protein complex prediction by date hub removal. Comput Biol Chem 74:407–419. https://doi.org/10.1016/j.compbiolchem.2018.03.012
Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D (2000) DIP: the database of interacting proteins. Nucleic Acids Res 28(1):289–291. https://doi.org/10.1093/nar/28.1.289
Rives AW, Galitski T (2003) Modular organization of cellular networks. Proc Natl Acad Sci 100(3):1128–1133. https://doi.org/10.1073/pnas.0237338100
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4(1):1–27. https://doi.org/10.1186/1471-2105-4-2
Palla G, Derényi I, Farkas I, Vicsek T (2005) Uncovering the overlapping community structure of complex networks in nature and society. Nature 435(7043):814–818. https://doi.org/10.1038/nature03607
Altaf-Ul-Amin M, Shinbo Y, Mihara K, Kurokawa K, Kanaya S (2006) Development and implementation of an algorithm for detection of protein complexes in large interaction networks. BMC Bioinform 7(1):1–13. https://doi.org/10.1186/1471-2105-7-207
Li XL, Foo CS, Ng SK (2007) Discovering protein complexes in dense reliable neighborhoods of protein interaction networks. Comput Syst Bioinform 6:157–168. https://doi.org/10.1142/9781860948732_0019
Wu M, Li X, Kwoh CK, Ng SK (2009) A core-attachment based method to detect protein complexes in PPI networks. BMC Bioinform 10(1):1–16. https://doi.org/10.1186/1471-2105-10-169
Nepusz T, Yu H, Paccanaro A (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9(5):471–472. https://doi.org/10.1038/nmeth.1938
Haque M, Sarmah R, Bhattacharyya DK (2018) A common neighbor based technique to detect protein complexes in PPI networks. J Genet Eng Biotechnol 16(1):227–238. https://doi.org/10.1016/j.jgeb.2017.10.010
Xiao Q, Luo P, Li M, Wang J, Wu FX (2019) A novel core-attachment-based method to identify dynamic protein complexes based on gene expression profiles and PPI networks. Proteomics 19(5):e1800129. https://doi.org/10.1002/pmic.201800129
Meng X, Xiang J, Zheng R, Wu FX, Li M (2021) DPCMNE: detecting protein complexes from protein-protein interaction networks via multi-level network embedding. IEEE/ACM Trans Comput Biol Bioinf 19(3):1592–602. https://doi.org/10.1109/TCBB.2021.3050102
King AD, Przulj N, Jurisica I (2004) Protein complex prediction via cost-based clustering. Bioinformatics 20(17):3013–3020. https://doi.org/10.1093/bioinformatics/bth351
Kovács IA, Palotai R, Szalay MS, Csermely P (2010) Community landscapes: an integrative approach to determine overlapping network module hierarchy, identify key nodes and predict network dynamics. PLoS One 5(9):e12528. https://doi.org/10.1371/journal.pone.0012528
Ou-Yang L, Yan H, Zhang XF (2017) A multi-network clustering method for detecting protein complexes from multiple heterogeneous networks. BMC Bioinform 18(13):23–34. https://doi.org/10.1186/s12859-017-1877-4
Wang J, Liang J, Zheng W, Zhao X, Mu J (2019) Protein complex detection algorithm based on multiple topological characteristics in PPI networks. Inf Sci 489:78–92. https://doi.org/10.1016/j.ins.2019.03.015
Enright AJ, Van Dongen S, Ouzounis CA (2002) An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res 30(7):1575–1584. https://doi.org/10.1093/nar/30.7.1575
Hwang W, Cho YR, Zhang A, Ramanathan M (2006) A novel functional module detection algorithm for protein-protein interaction networks. Algorithms Mol Biol 1(1):1–11. https://doi.org/10.1186/1748-7188-1-24
Peng W, Wang J, Zhao B, Wang L (2014) Identification of protein complexes using weighted pagerank-nibble algorithm and core-attachment structure. IEEE/ACM Trans Comput Biol Bioinf 12(1):179–192. https://doi.org/10.1109/TCBB.2014.2343954
Farutin V, Robison K, Lightcap E, Dancik V, Ruttenberg A, Letovsky S, Pradines J (2006) Edge-count probabilities for the identification of local protein communities and their organization. Proteins Struct Funct Bioinform 62(3):800–818. https://doi.org/10.1002/prot.20799
Ahn YY, Bagrow JP, Lehmann S (2010) Link communities reveal multiscale complexity in networks. Nature 466(7307):761–764. https://doi.org/10.1038/nature09182
Tasgin M, Herdagdelen A, Bingol H (2007) Community detection in complex networks using genetic algorithms. arXiv preprint arXiv:0711.0491. https://doi.org/10.48550/arXiv.0711.0491
Xu Y, Zhou J, Zhou S, Guan J (2017) CPredictor3.0: detecting protein complexes from PPI networks with expression data and functional annotations. BMC Syst Biol 11(7):45–56. https://doi.org/10.1186/s12918-017-0504-3
Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D (2004) The database of interacting proteins: 2004 update. Nucleic Acids Res 32:D449–D451. https://doi.org/10.1093/nar/gkh086
Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O’Donnell L, Leung G, McAdam R, Zhang F (2019) The BioGRID interaction database: 2019 update. Nucleic Acids Res 47(D1):D529–D541. https://doi.org/10.1093/nar/gky1079
Pu S, Wong J, Turner B, Cho E, Wodak SJ (2009) Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res 37(3):825–831. https://doi.org/10.1093/nar/gkn1005
Giurgiu M, Reinhard J, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Ruepp A (2019) CORUM: the comprehensive resource of mammalian protein complexes-2019. Nucleic Acids Res 47(D1):D559–D563. https://doi.org/10.1093/nar/gky973
Yamasaki C, Murakami K, Takeda JI, Sato Y, Noda A, Sakate R, Habara T, Nakaoka H, Todokoro F, Matsuya A, Imanishi T (2010) H-InvDB in 2009: extended database and data mining resources for human genes and transcripts. Nucleic Acids Res 38:D626–D632. https://doi.org/10.1093/nar/gkp1020
Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G (2004) GO: TermFinder-open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 20(18):3710–3715. https://doi.org/10.1093/bioinformatics/bth456
Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, Weng S (1998) SGD: Saccharomyces genome database. Nucleic Acids Res 26(1):73–79. https://doi.org/10.1093/nar/26.1.73
Pomaznoy M, Ha B, Peters B (2018) GOnet: a tool for interactive Gene Ontology analysis. BMC Bioinform 19(1):1–8. https://doi.org/10.1186/s12859-018-2533-3
Acknowledgements
We acknowledge the infrastructure and computational facilities received from DST-FIST Bioinformatics Lab of IIIT Bhubaneswar.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author confirms that there are no conflicts of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sahoo, T.R., Vipsita, S. & Patra, S. Complex Prediction in Large PPI Networks Using Expansion and Stripe of Core Cliques. Interdiscip Sci Comput Life Sci 15, 331–348 (2023). https://doi.org/10.1007/s12539-022-00541-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-022-00541-z