Abstract
Protein subcellular localization prediction is an important research area in bioinformatics, which plays an essential role in understanding protein function and mechanism. Many machine learning and deep learning algorithms have been employed for this task, but most of them do not use structural information of proteins. With the advances in protein structure research in recent years, protein contact map prediction has been dramatically enhanced. In this paper, we present GraphLoc, a deep learning model that predicts the localization of proteins at the subcellular level. The cores of the model are a graph convolutional neural network module and a multi-head attention module. The protein topology graph is constructed based on a contact map predicted from protein sequences, which is used as the input of the GCN module to take full advantage of the structural information of proteins. Multi-head attention module learns the weighted contribution of different amino acids to subcellular localization in different feature representation subspaces. Experiments on the benchmark dataset show that the performance of our model is better than others. The code can be accessed at https://github.com/GoodGuy398/GraphLoc.
Graphical abstract
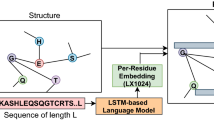
The proposed GraphLoc model consists of three parts. The first part is a graph convolutional network (GCN) module, which utilizes the predicted contact maps to construct protein graph, taking benefit of protein information accordingly. The second part is the multi-head attention module, which learns the weighted contribution of different amino acids in different feature representation subspace, and weighted average the feature map across all amino acid nodes. The last part is a fully connected layer that maps the flatten graph representation vector to another vector with a category number dimension, followed by a softmax layer to predict the protein subcellular localization.





Similar content being viewed by others
References
Davis JR, Kakar M, Lim CS (2007) Controlling protein compartmentalization to overcome disease. Pharm Res 24(1):17–27. https://doi.org/10.1007/s11095-006-9133-z
Hung MC, Link W (2011) Protein localization in disease and therapy. J Cell Sci 124(20):3381–3392. https://doi.org/10.1242/jcs.089110
Walther TC, Mann M (2010) Mass spectrometry–based proteomics in cell biology. J Cell Biol 190(4):491–500. https://doi.org/10.1083/jcb.201004052
Schubert W, Bonnekoh B, Pommer AJ, Philipsen L, Bockelmann R, Malykh Y, Gollnick H, Friedenberger M, Bode M, Dress AWM (2006) Analyzing proteome topology and function by automated multidimensional fluorescence microscopy. Nat Biotechnol 24(10):1270–1278. https://doi.org/10.1038/nbt1250
Imai K, Nakai K (2010) Prediction of subcellular locations of proteins: where to proceed? Proteomics 10(22):3970–3983. https://doi.org/10.1002/pmic.201000274
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT et al (2000) Gene ontology: tool for the unification of biology. Nat Genet 25(1):25–29. https://doi.org/10.1038/75556
Kosugi S, Hasebe M, Tomita M, Yanagawa H (2008) Nuclear export signal consensus sequences defined using a localization-based yeast selection system. Traffic 9(12):2053–2062. https://doi.org/10.1111/j.1600-0854.2008.00825.x
Sperschneider J, Catanzariti AM, DeBoer K, Petre B, Gardiner DM, Singh KB, Dodds PN (2017) Taylor JM (2017) Localizer: subcellular localization prediction of both plant and effector proteins in the plant cell. Sci Rep 7(1):1–14. https://doi.org/10.1038/srep44598
Blum T, Briesemeister S, Kohlbacher O (2009) Multiloc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinformatics 10(1):1–11. https://doi.org/10.1186/1471-2105-10-274
Briesemeister S, Rahnenfhrer J, Kohlbacher O (2010) Yloc—an interpretable web server for predicting subcellular localization. Nucleic Acids Res 38(suppl 2):W497–W502. https://doi.org/10.1093/nar/gkq477
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Collier CJA, Nakai K (2007) Wolf psort: protein localization predictor. Nucleic Acids Res 35(suppl 2):W585–W587. https://doi.org/10.1093/nar/gkm259
Du L, Meng Q, Chen Y, Wu P (2020) Subcellular location prediction of apoptosis proteins using two novel feature extraction methods based on evolutionary information and lda. BMC Bioinformatics 21(1):1–19. https://doi.org/10.1186/s12859-020-3539-1
Yadav AK, Singla D (2020) Vacpred: Sequence-based prediction of plant vacuole proteins using machine-learning techniques. J Biosci 45(1):1–9. https://doi.org/10.1007/s12038-020-00076-9
Savojardo C, Martelli PL, Fariselli P, Casadio R (2015) Tppred3 detects and discriminates mitochondrial and chloroplastic targeting peptides in eukaryotic proteins. Bioinformatics 31(20):3269–3275. https://doi.org/10.1093/bioinformatics/btv367
Small I, Peeters N, Legeai F, Lurin C (2004) Predotar: a tool for rapidly screening proteomes for n-terminal targeting sequences. Proteomics 4(6):1581–1590. https://doi.org/10.1002/pmic.200300776
Emanuelsson O, Brunak S, Heijne GV, Nielsen H (2007) Locating proteins in the cell using targetp, signalp and related tools. Nat Protoc 2(4):953–971. https://doi.org/10.1038/nprot.2007.131
Armenteros JJA, Sønderby CK, Sønderby SK, Nielsen H, Winther O (2017) Deeploc: prediction of protein subcellular localization using deep learning. Bioinformatics 33(21):3387–3395. https://doi.org/10.1093/bioinformatics/btx431
Long W, Yang Y, Shen HB (2020) Imploc: a multi-instance deep learning model for the prediction of protein subcellular localization based on immunohistochemistry images. Bioinformatics 36(7):2244–2250. https://doi.org/10.1093/bioinformatics/btz909
Jiang Y, Wang D, Yao Y, Eubel H, Kunzler P, Møller IM, Xu D (2021) Mulocdeep: a deep-learning framework for protein subcellular and suborganellar localization prediction with residue-level interpretation. Comput Struct Biotechnol J 19:4825–4839. https://doi.org/10.1016/j.csbj.2021.08.027
Cong H, Liu H, Cao Y, Chen Y, Liang C (2022) Multiple protein subcellular locations prediction based on deep convolutional neural networks with self-attention mechanism. Interdiscip Sci Comput Life Sci 1–18. https://doi.org/10.1007/s12539-021-00496-7
Gligorijevic V, Barot M, Bonneau R (2018) deepnf: deep network fusion for protein function prediction. Bioinformatics 34(22):3873–3881. https://doi.org/10.1093/bioinformatics/bty440
Schaarschmidt J, Monastyrskyy B, Kryshtafovych A, Bonvin AMJ (2018) Assessment of contact predictions in casp12: co-evolution and deep learning coming of age. Proteins Struct Funct Bioinf 86:51–66. https://doi.org/10.1002/prot.25407
Hanson J, Paliwal K, Litfin T, Yang Y, Zhou Y (2018) Accurate prediction of protein contact maps by coupling residual two-dimensional bidirectional long short-term memory with convolutional neural networks. Bioinformatics 34(23):4039–4045. https://doi.org/10.1093/bioinformatics/bty481
Kipf TN, Welling M (2016) Semi-supervised classification with graph convolutional networks. arXiv:1609.02907. https://doi.org/10.48550/arXiv.1609.02907
Mount DA (2008) Using blosum in sequence alignments. Cold Spring Harbor Protocols (6):pdb–top39. https://doi.org/10.1101/pdb.top39
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22(13):1658–1659. https://doi.org/10.1093/bioinformatics/btl158
Bairoch A, Apweiler R (2000) The swiss-prot protein sequence database and its supplement trembl in 2000. Nucleic Acids Res 28(1):45–48. https://doi.org/10.1093/nar/28.1.45
Venkatarajan MS, Braun W (2001) New quantitative descriptors of amino acids based on multidimensional scaling of a large number of physical–chemical properties. Mol Model Annu 7(12):445–453. https://doi.org/10.1007/s00894-001-0058-5
Emerson IS, Amala A (2017) Protein contact maps: a binary depiction of protein 3d structures. Phys A 465:782–791. https://doi.org/10.1016/j.physa.2016.08.033
Zheng S, Yan X, Yang Y, Xu J (2019) Identifying structure–property relationships through smiles syntax analysis with self-attention mechanism. J Chem Inf Model 59(2):914–923. https://doi.org/10.1021/acs.jcim.8b00803
Lin Z, Feng M, Nogueira dos Santos C, Yu M, Xiang B, Zhou B, Bengio Y (2017) A structured self-attentive sentence embedding. arXiv preprint arXiv:1703.03130. https://doi.org/10.48550/arXiv.1703.03130
Yang H, Wang M, Yu Z, Zhao XM, Li A (2020) Gancon: Protein contact map prediction with deep generative adversarial network. IEEE Access 8:80899–80907. https://doi.org/10.1109/ACCESS.2020.2991605
Acknowledgements
This research was funded by the National Natural Science Foundation of China (61972174), Key research and development project of Jilin Provincial Science and Technology Department (20210201080GX), Jilin Province Development and Reform Commission (2021C044-1), Guangdong science and technology planning (2020A0505100018), Guangdong universities’ innovation team (2021KCXTDO15), and Guangdong key disciplines (2021ZDJS138) projects.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
About this article
Cite this article
Zhang, T., Gu, J., Wang, Z. et al. Protein Subcellular Localization Prediction Model Based on Graph Convolutional Network. Interdiscip Sci Comput Life Sci 14, 937–946 (2022). https://doi.org/10.1007/s12539-022-00529-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-022-00529-9