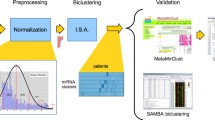
Abstract
The influence of non-coding RNAs, such as lncRNAs (long non-coding RNAs) and miRNAs (microRNAs), is undeniable in several diseases, for example, in the formation of neoplasms and cancer scenarios. However, there are challenges due to the scarcity of validated datasets and the imbalance in the data. We found that the research of associations between miRNAs-lncRNAs and diseases is limited or done separately. In addition, those investigations, which use Machine Learning models joined with genomic sequence features extracted from miRNAs and lncRNAs, are few compared with using some methods such as genomic expression or Deep Learning techniques. In this paper, we propose a structure of using supervised and unsupervised machine learning models with genomic sequence features, such as k-mers, sequence alignments, and energy folding values, to validate miRNAs and lncRNAs association with breast cancer and neoplasms scenarios. Using One-Class SVM for outlier detection and comparing two supervised models such as SVM and Random Forest, we manage to obtain accuracy results of 95.44% for the One-class model, with 88.79% and 99.65% for the SVM and Random Forest models, respectively. The results showed a promising path for the study of sequence features interactions joined with Machine Learning models comparable to those found in the existing literature.
Graphic Abstract






Similar content being viewed by others
References
Coleman WB (2020) Neoplasia. Essential concepts in molecular pathology. Elsevier, Amsterdam, pp 55–80
Vogelstein B, Kinzler K (2002) Genetic basis of human cancer, 2nd edn. McGraw-Hill, New York, p 28
Harries LW (2012) Long non-coding RNAs and human disease. Biochem Soc Trans 40:902–906. https://doi.org/10.1042/BST20120020
Wapinski O, Chang HY (2011) Long noncoding RNAs and human disease. Trends Cell Biol 21:354–361. https://doi.org/10.1016/j.tcb.2011.04.001
Loh H-Y, Norman BP, Lai K-S et al (2019) The regulatory role of MicroRNAs in breast cancer. IJMS 20:4940. https://doi.org/10.3390/ijms20194940
McAnena P, Tanriverdi K, Curran C et al (2019) Circulating microRNAs miR-331 and miR-195 differentiate local luminal a from metastatic breast cancer. BMC Cancer 19:436. https://doi.org/10.1186/s12885-019-5636-y
Fu L, Peng Q (2017) A deep ensemble model to predict miRNA-disease association. Sci Rep 7:14482. https://doi.org/10.1038/s41598-017-15235-6
Guo Z-H, You Z-H, Wang Y-B et al (2019) A learning-based method for LncRNA-disease association identification combing similarity information and rotation forest. iScience 19:786–795. https://doi.org/10.1016/j.isci.2019.08.030
Huang Y-A, Huang Z-A, You Z-H et al (2019) Predicting lncRNA-miRNA Interaction via Graph Convolution Auto-Encoder. Front Genet 10:758. https://doi.org/10.3389/fgene.2019.00758
Wen J, Liu Y, Shi Y et al (2019) A classification model for lncRNA and mRNA based on k-mers and a convolutional neural network. BMC Bioinform 20:469. https://doi.org/10.1186/s12859-019-3039-3
Miao Y-R, Liu W, Zhang Q, Guo A-Y (2018) lncRNASNP2: an updated database of functional SNPs and mutations in human and mouse lncRNAs. Nucleic Acids Res 46:D276–D280. https://doi.org/10.1093/nar/gkx1004
Zhao Y, Li H, Fang S et al (2016) NONCODE 2016: an informative and valuable data source of long non-coding RNAs. Nucleic Acids Res 44:D203–D208. https://doi.org/10.1093/nar/gkv1252
Lu M, Shi B, Wang J et al (2010) TAM: A method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinform 11:419. https://doi.org/10.1186/1471-2105-11-419
Xu J, Wong C-W (2013) Enrichment analysis of miRNA targets. In: Ying S-Y (ed) MicroRNA protocols. Humana Press, Totowa, pp 91–103
Rehman O, Zhuang H, Muhamed Ali A et al (2019) Validation of miRNAs as breast cancer biomarkers with a machine learning approach. Cancers 11:431. https://doi.org/10.3390/cancers11030431
Zhang P, Meng J, Luan Y, Liu C (2020) Plant miRNA–lncRNA interaction prediction with the ensemble of CNN and IndRNN. Interdiscip Sci Comput Life Sci 12:82–89. https://doi.org/10.1007/s12539-019-00351-w
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucl Acids Res 42:D68–D73. https://doi.org/10.1093/nar/gkt1181
Tan P-N, Steinbach M, Kumar V (2005) Introduction to data mining, 1st edn. Addison-Wesley Longman Publishing Co., Inc., Boston
Hofacker IL (2003) Vienna RNA secondary structure server. Nucleic Acids Res 31:3429–3431. https://doi.org/10.1093/nar/gkg599
Yang S, Wang Y, Lin Y et al (2020) LncMirNet: predicting LncRNA–miRNA interaction based on deep learning of ribonucleic acid sequences. Molecules 25:4372. https://doi.org/10.3390/molecules25194372
Acknowledgements
This research is supported partially by South African National Research Foundation Grants (Nos. 114911 & 132797) and Tertiary Education Support Programme (TESP) of South African ESKOM.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gutiérrez-Cárdenas, J., Wang, Z. Classification of Breast Cancer and Breast Neoplasm Scenarios Based on Machine Learning and Sequence Features from lncRNAs–miRNAs-Diseases Associations. Interdiscip Sci Comput Life Sci 13, 572–581 (2021). https://doi.org/10.1007/s12539-021-00451-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00451-6