Abstract
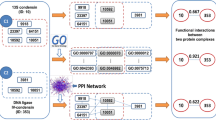
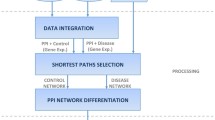
One important challenge in the post-genomic era is to explore disease mechanisms by efficiently integrating different types of biological data. In fact, a single disease is usually caused through multiple genes products such as protein complexes rather than single gene. Therefore, it is meaningful for us to discover protein communities from the protein–protein interaction network and use them for inferring disease–disease associations. In this article, we propose a new framework including protein–protein networks, disease–gene associations and disease–complex pairs to cluster protein complexes and infer disease associations. Complexes discovered by our approach is superior in quality (Sn, PPV and ACC) and clustering quantity than other four popular methods on three PPI networks. A systematic analysis shows that disease pairs sharing more protein complexes (such as Glucose and Lipid Metabolic Disorders) are more similar and overlapping proteins may have different roles in different diseases. These findings can provide clinical scholars and medical practitioners with new ideas on disease identification and treatment.






Similar content being viewed by others
References
Barabasi AT, Gulbahce N, Loscalzo J (2011) Network medicine: a network-based approach to human disease. Nat Rev Genet 12:56–68. https://doi.org/10.1038/nrg2918
Zhuang Q, An X, Liu H et al (2019) Uncovering the resistance mechanism of mycobacterium tuberculosis to rifampicin due to RNA polymerase H451D/Y/R mutations from computational perspective. Front Chem 7:819. https://doi.org/10.3389/fchem.2019.00819
Li ZC, Huang QX, Chen XY et al (2020) Identification of drug-disease associations using information of molecular structures and clinical symptoms via deep convolutional neural network. Front Chem 7:924. https://doi.org/10.3389/fchem.2019.00924
Hu L, Chan KCC (2015) A density-based clustering approach for identifying overlapping protein complexes with functional preferences. BMC Bioinf 16:174. https://doi.org/10.1186/s12859-015-0583-3
Tang XQ, Zhu P (2013) Hierarchical clustering problems and analysis of fuzzy proximity relation on granular space. IEEE Trans Fuzzy Syst 21:814–824. https://doi.org/10.1109/TFUZZ.2012.2230176
Maddi AMA, Moughari FA, Balouchi M et al (2019) CDAP: an online package for evaluation of complex detection methods. Sci Rep 9:12751. https://doi.org/10.1038/s41598-019-49225-7
Doorbar J, Egawa N, Griffin H et al (2015) Human papillomavirus molecular biology and disease association. Rev Med Virol 25:2–23. https://doi.org/10.1002/rmv.1822
Shen XJ, Yi L, Jiang XP et al (2016) Neighbor affinity based algorithm for discovering temporal protein complex from dynamic PPI network. Methods 110:90–96. https://doi.org/10.1016/j.ymeth.2016.06.010
Ren J, Wang JX, Li M, Wu FX (2015) Discovering essential proteins based on PPI network and protein complex. Int J Data Min Bioinf 12:24–43. https://doi.org/10.1504/IJDMB.2015.068951
Ahn YY, Bagrow JP, Lehmann S (2010) Link communities reveal multiscale complexity in networks. Nature 466:761–764. https://doi.org/10.1038/nature09182
Wei G, Tao Z, Tao H, Cai YD (2020) Disease cluster detection and functional characterization. IEEE Access 99:1–1. https://doi.org/10.1109/ACCESS.2020.3013666
Bouguettaya A, Yu Q, Liu XM et al (2015) Efficient agglomerative hierarchical clustering. Exp Syst Appl 42:2785–2797. https://doi.org/10.1016/j.eswa.2014.09.054
Palla G, Derenyi I, Farkas I et al (2005) Uncovering the overlapping community structure of complex networks in nature and society. Nature 435:814–818. https://doi.org/10.1038/nature03607
Liu G, Wong L, Chua HN (2009) Complex discovery from weighted PPI networks. Bioinformatics 25:1891–1897. https://doi.org/10.1093/bioinformatics/btp311
Lancichinetti A, Fortunato S, Kertész J (2009) Detecting the overlapping and hierarchical community structure in complex networks. New J Phys 11:033015. https://doi.org/10.1088/1367-2630/11/3/033015
Nepusz T, Yu H, Paccanaro A (2012) Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods 9:471-U81. https://doi.org/10.1038/nmeth.1938
Maddi AMA, Eslahchi C (2017) Discovering overlapped protein complexes from weighted PPI networks by removing inter-module hubs. Sci Rep 7:3247. https://doi.org/10.1038/s41598-017-03268-w
Ding Z, Zhang X, Sun D et al (2016) Overlapping community detection based on network decomposition. Sci Rep 6:24115. https://doi.org/10.1038/srep24115
Karrer B, Newman MEJ (2010) Stochastic blockmodels and community structure in networks. Phys Rev E 83:016107. https://doi.org/10.1103/PhysRevE.83.016107
Gregory S (2010) Finding overlapping communities in networks by label propagation. New J Phys 12:103018. https://doi.org/10.1088/1367-2630/12/10/103018
Wen X, Chen WN, Lin Y et al (2017) A maximal clique based multiobjective evolutionary algorithm for overlapping community detection. IEEE Trans Evolut Comput 21:363–377. https://doi.org/10.1109/TEVC.2016.2605501
Batool Z, Usman M, Saleem K et al (2018) Disease-disease association using network modeling: challenges and opportunities. J Med Image Health Inf 8:627–638. https://doi.org/10.1166/jmihi.2018.2342
Qi JM, Zhou JX, Tang XQ, Wang YL (2020) Gene biomarkers derived from clinical data of hepatocellular carcinoma. Interdiscip Sci Comput Life Sci 12:226–236. https://doi.org/10.1007/s12539-020-00366-8
Higalgo CA, Blumm N, Barabasi AL et al (2009) A dynamic network approach for the study of human phenotypes. PLoS Comput Biol 5:e1000353. https://doi.org/10.1371/journal.pcbi.1000353
Gamba A, Salmona M, Bazzoni G (2020) Quantitative analysis of proteins which are members of the same protein complex but cause locus heterogeneity in disease. Sci Rep 10:10423. https://doi.org/10.1038/s41598-020-66836-7
Ni P, Wang JX, Zhong P et al (2018) Constructing disease similarity networks based on disease module theory. IEEE-ACM Trans Comput Biol Bioinf 17:906–915. https://doi.org/10.1109/TCBB.2018.2817624
Choobdar S, Ahsen ME, Crawford J et al (2019) Assessment of network module identification across complex diseases. Nat Methods 16:843–852. https://doi.org/10.1038/s41592-019-0509-5
Goh KI, Cusick ME, Valle D et al (2007) The human disease network. Proc Natl Acad Sci USA 104:8685–8690. https://doi.org/10.1073/pnas.0701361104
Wang QH, Liu WS, Ning SW et al (2012) Community of protein complexes impacts disease association. Eur J Genet 20:1162–1167. https://doi.org/10.1038/ejhg.2012.74
Szklarczyk D, Franceschini A, Kuhn M et al (2011) The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucl Acids Res 39:D561–D568. https://doi.org/10.1093/nar/gkq973
Giurgiu M, Reinhard J, Brauner B et al (2019) CORUM: the comprehensive resource of mammalian protein complexes 2019. Nucl Acids Res 47:D559–D563. https://doi.org/10.1093/nar/gky973
Pinero J, Bravo A, Queralt-Rosinach N et al (2017) DisGeNET: a comprehensive platform integrating information on human disease-associated genes and variants. Nucl Acids Res 45:D833–D839. https://doi.org/10.1093/nar/gkw943
Radicchi F, Castellano C et al (2004) Defining and identifying communities in networks. Proc Natl Acad Sci USA 101:2658–2663. https://doi.org/10.1073/pnas.0400054101
Leskovec J, Kleinberg J, Faloutsos C (2005) Graphs over time: densification laws, shrinking diameters and possible explanations. ACM. https://doi.org/10.1145/1081870.1081893
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinf 4:2. https://doi.org/10.1186/1471-2105-4-2
Brohee S, van-Helden J, (2006) Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinf 7:488. https://doi.org/10.1186/1471-2105-7-488
Jaccard P (1912) The distribution of the flora in the alpine zone. New Phytol 11:37–50. https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Chen LG, Chen XW, Huang X et al (2019) Regulation of glucose and lipid metabolism in health and disease. Sci Chin Life Sci 62:1420–1458. https://doi.org/10.1007/s11427-019-1563-3
Recalde D, Cenarro A, Garcia-Otin AL et al (2002) Analysis of apolipoprotein A-I, lecithin: cholesterol acyltransferase and glucocerebrosidase genes in hypoalphalipoproteinemia. Atherosclerosis 163:49–58. https://doi.org/10.1016/S0021-9150(01)00753-5
Yates JRW, Sepp T, Matharu BK et al (2007) Complement C3 variant and the risk of age-related macular degeneration. New Engl J Med 357:553–561. https://doi.org/10.1056/NEJMoa072618
Zhao Q, Zhang Y, Hu H et al (2018) (2018) IRWNRLPI: integrating random walk and neighborhood regularized logistic matrix factorization for lncRNA-protein interaction prediction. Front Genet 9:239. https://doi.org/10.3389/fgene.2018.00239.eCollection
Hu H, Zhang L, Ai HX et al (2018) HLPI-ensemble: prediction of human lncRNA-protein interactions based on ensemble strategy. RNA Biol 15:797–806. https://doi.org/10.1080/15476286.2018.1457935
Zhao Q, Yu HF, Ming Z et al (2018) The bipartite network projection-recommended algorithm for predicting long non-coding RNA-protein interactions. Mol Therap Nucl Acids 13:464–471. https://doi.org/10.1016/j.omtn.2018.09.020
Funding
This work has been supported by the National Natural Science Foundation of China (Grand No. 11371174).
Author information
Authors and Affiliations
Contributions
XQT and LX designed the study. LX implemented the analysis. LX and XQT wrote the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflicts of interest in this paper.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xue, L., Tang, XQ. A New Framework for Discovering Protein Complex and Disease Association via Mining Multiple Databases. Interdiscip Sci Comput Life Sci 13, 683–692 (2021). https://doi.org/10.1007/s12539-021-00432-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-021-00432-9