Abstract
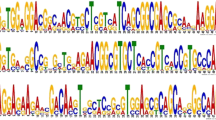
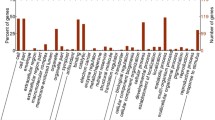
Cereal grain bread wheat (T. aestivum) is an important source of food and belongs to Poaceae family. Hypothetical proteins (HPs), i.e., proteins with unknown functions, share a substantial portion of wheat proteomes and play important roles in growth and physiology of plant system. Several functional annotations studies utilizing the protein sequences for characterization of role of individual protein in physiology of plant systems were being reported in recent past. In this study, an integrated pipeline of software/servers has been used for the identification and functional annotation of 124 unique HPs of T. aestivum considering available data in NCBI till date. All HPs were broadly annotated, out of which functions of 77 HPs were successfully assigned with high confidence level. Precisely functional annotation of remaining 47 HPs is also characterized with low confidence. Several latest versions of protein family databases, pathways information, genomics context methods and in silico tools were utilized to identify and assign function for individual HPs. Annotation result of several HPs mainly belongs to cellular protein, metabolic enzymes, binding proteins, transmembrane proteins, transcription factors and photosystem regulator proteins. Subsequently, functional analysis has revealed the role of few HPs in abiotic stress, which were further verified by phylogenetic analysis. The functionally associated proteins with each of above-mentioned abiotic stress-related proteins were identified through protein–protein interaction network analysis. The outcome of this study may be helpful for formulating general set pipeline/protocols for a better understanding of the role of HPs in physiological development of various plant systems.






Similar content being viewed by others
References
Curtis BC (2002) Wheat in the world. Bread wheat: improvement and production (No. CIS-3616. CIMMYT)
Padulosi S, Hammer K, Heller J (1996) Hulled wheats. Promoting the conservation and use of underutilized and neglected crop 4. In: Proceeding of the first international workshop on Hulled, Tuscany (Italia), 21–22 Jul 1995. IPGRI, Roma
Eversole K, Feuillet C, Mayer KF, Rogers J (2014) Slicing the wheat genome. Science 345:285–287
Mayer KF, Rogers J, Doležel J, Pozniak C, Eversole K, Feuillet C, Ayling S (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345:1251788
Galperin MY, Koonin EV (2004) Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucl Acids Res 32:5452–5463
Brenchley R, Spannagl M, Pfeifer M, Barker GL, D`Amore R, Allen AM, Hall N (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491:705–710
Zarembinski TI, Hung LW, Mueller-Dieckmann HJ, Kim KK, Yokota H, Kim R, Kim SH (1998) Structure-based assignment of the biochemical function of a hypothetical protein: a test case of structural genomics. Proc Natl Acad Sci 95:15189–15193
Doerks T, von Mering C, Bork P (2004) Functional clues for hypothetical proteins based on genomic context analysis in prokaryotes. Nucl Acids Res 32:6321–6326
Desler C, Suravajhala P, Sanderhoff M, Rasmussen LJ (2009) In silico screening for functional candidates amongst hypothetical proteins. BMC Bioinform 10:289
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucl Acids Res 31:3784–3788
Blum T, Briesemeister S, Kohlbacher O (2009) MultiLoc2: integrating phylogeny and gene ontology terms improves subcellular protein localization prediction. BMC Bioinform 10:274
Briesemeister S, Blum T, Brady S, Lam Y, Kohlbacher O, Shatkay H (2009) SherLoc2: a high-accuracy hybrid method for predicting subcellular localization of proteins. J Prot Res 8:5363–5366
Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, Nakai K (2007) WoLF PSORT: protein localization predictor. Nucl Acids Res 35:W585–W587
Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using targetP, signalP and related tools. Nat Protoc 2:953–971
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucl Acids Res 33:W244–W248
Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Sonnhammer EL (2013) Pfam: the protein families database. Nucl Acids Res 42:D222–D230
Sillitoe I, Cuff AL, Dessailly BH, Dawson NL, Furnham N, Lee D, Orengo CA (2012) New functional families (FunFams) in CATH to improve the mapping of conserved functional sites to 3D structures. Nucl Acids Res 41:D490–D498. doi:10.1093/nar/gks1211
Geer LY, Domrachev M, Lipman DJ, Bryant SH (2002) CDART: protein homology by domain architecture. Genome Res 12:1619–1623
Conte LL, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C (2000) SCOP: a structural classification of proteins database. Nucl Acids Res 28:257–259
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucl Acids Res 40:D302–D305
Rost B, Valencia A (1996) Pitfalls of protein sequence analysis. Curr Opin Biotechnol 7:457–461
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, Martin MJ (2004) UniProt: the universal protein knowledge base. Nucl Acids Res 32:D115–D119
Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Brice MD, Rodgers JR, Tasumi M (1978) The protein data bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys 185:584–591
Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Bryant SH (2011) CDD: a conserved domain database for the functional annotation of proteins. Nucl Acids Res 39:D225–D229
Krogh A, Larsson B, Von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580
Rappoport N, Karsenty S, Stern A, Linial N, Linial M (2011) ProtoNet 6.0: organizing 10 million protein sequences in a compact hierarchical family tree. Nucl Acids Res 40:D313–D320. doi:10.1093/nar/gkr1027
Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Schneider M (2003) The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucl Acids Res 31:365–370
Bairoch A, Apweiler R (1999) The SWISS-PROT protein sequence data bank and its supplement TrEMBL in 1999. Nucl Acids Res 27:49–54
Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Searle SM (2013) Ensembl 2013. Nucl Acids Res 41:D48–D55
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, von Mering C (2014) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucl Acids Res 43:D447–D452
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mole Biol Evol 30:2725–2729
Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ (2009) Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucl Acids Res 22:4673–4680
Cozzone AJ (2010) Proteins: fundamental chemical properties. eLS. doi:10.1002/9780470015902.a0001330.pub2
Sturm A (1999) Invertases, primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiol 121:1–8
Minic Z (2008) Physiological roles of plant glycoside hydrolases. Planta 227:723–740
Dramé KN, Passaquet C, Repellin A, Zuily-Fodil Y (2013) Cloning, characterization and differential expression of a Bowman-Birk inhibitor during progressive water deficit and subsequent recovery in peanut (Arachis hypogaea) leaves. J Plant Physiol 170:225–229
Prasad CVS, Gupta S, Gaponenko A, Dhar M (2012) In-silico comparative study of inhibitory mechanism of plant serine proteinase inhibitors. Bioinformation 8:673–678
Lindahl T, Ljungquist S, Siegert W, Nyberg B, Sperens BDNA (1977) DNA N-glycosidases: properties of uracil-DNA glycosidase from Escherichia coli. J Biol Chem 252:3286–3294
D’Auria JC, Reichelt M, Luck K, Svatoš A, Gershenzon J (2007) Identification and characterization of the BAHD acyltransferase malonyl CoA: anthocyanidin 5-O-glucoside-6″-O-malonyltransferase (At5MAT) in Arabidopsis thaliana. FEBS Lett 581:872–878
Treimer JF, Zenk MH (1979) Purification and properties of strictosidine synthase, the key enzyme in indole alkaloid formation. Eur J Biochem 101:225–233
Akoh CC, Lee GC, Liaw YC, Huang TH, Shaw JF (2004) GDSL family of serine esterases/lipases. Prog Lipid Res 43:534–552
Sanchez R, Zhou MM (2011) The PHD finger: a versatile epigenome reader. Trends Biochem Sci 36:364–372
Ortega-Galisteo AP, Morales-Ruiz T, Ariza RR, Roldán-Arjona T (2008) Arabidopsis DEMETER-LIKE proteins DML2 and DML3 are required for appropriate distribution of DNA methylation marks. Plant Mol Biol 67:671–681
Zhao Q, Leung S, Corbett AH, Meier I (2006) Identification and characterization of the Arabidopsis orthologs of nuclear transport factor 2, the nuclear import factor of ran. Plant Physiol 140:869–878
Miyakawa T, Hatano KI, Miyauchi Y, Suwa YI, Sawano Y, Tanokura M (2014) A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol 166:766–787
Canel C, Bailey-Serres JN, Roose ML (1995) Pummelo fruit transcript homologous to ripening-induced genes. Plant Physiol 108:1323–1324
Padmanabhan V, Dias DM, Newton RJ (1997) Expression analysis of a gene family in loblolly pine (Pinus taeda L.) induced by water deficit stress. Plant Mol Biol 35:801–807
Guo WJ, Ho THD (2008) An abscisic acid-induced protein, HVA22, inhibits gibberellin-mediated programmed cell death in cereal aleurone cells. Plant Physiol 147:1710–1722
Baulcombe D, Lazarus C, Martienssen R (1984) Gibberellins and gene control in cereal aleurone cells. J Embryol Exp Morphol 83:119–135
Hong-Bo S, Zong-Suo L, Ming-An S (2005) LEA proteins in higher plants: structure, function, gene expression and regulation. Colloids Surf B Biointerf 45:131–135
Scanlon MJ, Norton RS (1994) Multiple conformations of the sea anemone polypeptide anthopleurin-A in solution. Protein Sci 3:1121–1124
Komatsu S (2008) Plasma membrane proteome in Arabidopsis and rice. Proteomics 8:4137–4145
Ebert JC, Altman RB (2008) Robust recognition of zinc binding sites in proteins. Protein Sci 17:54–65
Ruijter ND, Emons AMC (1999) Actin-binding proteins in plant cells. Plant Biol 1:26–35
Chinnusamy V, Gong Z, Zhu JK (2008) Nuclear RNA export and its importance in abiotic stress responses of plants. In nuclear pre-mRNA processing in plants. Springer, Berlin, pp 235–255
Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K (2005) Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure 13:143–153
Naver H, Boudreau E, Rochaix JD (2001) Functional studies of ycf3 its role in assembly of photosystem I and interactions with some of its subunits. Plant Cell 13:2731–2745
Lin R, Ding L, Casola C, Ripoll DR, Feschotte C, Wang H (2007) Transposase-derived transcription factors regulate light signaling in Arabidopsis. Science 318:1302–1305
Busby S, Ebright RH (1999) Transcription activation by catabolite activator protein (CAP). J Mol Biol 293:199–213
Imai K, Nakai K (2010) Prediction of subcellular locations of proteins: where to proceed? Proteomics 10:3970–3983
Shahbaaz M, Imtaiyaz Hassan M, Ahmad F (2013) Functional annotation of conserved hypothetical proteins from Haemophilus influenzae Rd KW20. PLoS One 8:e84263
Naqvi AAT, Ahmad F, Hassan MI (2015) Identification of functional candidates amongst hypothetical proteins of Mycobacterium leprae BR4923, a causative agent of leprosy. Genome 58:25–42
Acknowledgments
Authors are thankful to the Indian Institute of Information Technology, Allahabad, for providing the required infrastructure and computational facilities to complete this work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Funding Information
This study was not supported by any funding agency.
Human rights and animal statement
This research does not perform any experiment on human and animals. All data used in this in silico work collected from the open sources. Hence, authors declare that there is no compliance with ethical standards.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gupta, S., Singh, Y., Kumar, H. et al. Identification of Novel Abiotic Stress Proteins in Triticum aestivum Through Functional Annotation of Hypothetical Proteins. Interdiscip Sci Comput Life Sci 10, 205–220 (2018). https://doi.org/10.1007/s12539-016-0178-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-016-0178-3