Abstract
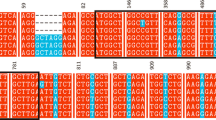
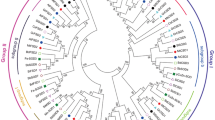
Superoxide dismutases are a class of enzymes that catalyze the dismutation of superoxide into oxygen and hydrogen peroxide. As such, they are an important antioxidant defense in nearly all cells exposed to oxygen. Superoxide dismutase (SOD) acts as first line of defense against oxidative and genetic stress. Manganese superoxide dismutase (MnSOD), found in mitochondria or peroxisomes, contains Mn (III) at the active site. The three dimensional structure of MnSOD of Oryza sativa is not yet available in protein data bank so we have predicted the structure model of O. sativa MnSOD using homology modeling. The predicted model can further be explored for identification of ligand binding sites which may be useful for understanding specific role in functional site residues during catalysis. This study also demonstrated that the phylogenetic analysis of O. sativa MnSOD protein with distinct dicot and monocot plant species. The MnSOD protein of O. sativa has shown similarity with both monocot and as well as dicot plant species.
Similar content being viewed by others
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl Acids Res 25: 3389–3402.
Arnold, K., Bordoli, L., Kopp, J., Schwede, T. 2006. The SWISS-MODEL workspace: a web-based environ ment for protein structure homology modeling. Bioinformatics 22: 195–201.
Brady, G., and Stouten, P. 2000. Fast prediction and visualization of protein binding pockets with PASS. J Comput Aided Mol Des 14: 383–401.
Bowler, C., Alliotte, T., Van Den Bulcke, M., Bauw, G., Vandekerckhove, J., Van Montagu, M., Inze, D. 1989. A plant manganese superoxide dismutase is efficiently imported and correctly processed by yeast mitochondria. Proc. Natl. Acad. Sci. U.S.A. 86: 3237–3241.
Bowler, C., Slooten, L., Vandenbranden, S., De Rycke, R., Botterman, J., Sybesma, C., Van Montagu, M., Inze, D. 1991. Manganese superoxide dismutase can reduce cellular damage mediated by oxygen radicals in transgenic plants. EMBO J, 10(7): 1723–1732.
Bowler, C., Van Montagu, M., Inze, D. 1992. Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 43: 83–116.
Berman, H.M., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T.N., Weissig, H., Shindyalov, I. N., Bourne, P. E. 2000. The Protein Data Bank. Nucleic Acids Research 28: 235–242.
DeLano, W.L. 2002. The PyMOL Molecular Graphics System. DeLano Scientific, San Carlos, CA, USA.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39: 783–791.
Henikoff, S., Henikoff, J.G. 1992. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 89: 10915–10919.
Hess, B., Kutzner, C., van der Spoel, D., Lindahl, E. 2008. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J. Chem. Theory. Comput. 4: 435–447.
Laskowski, R.A., MacArthur, M.W., Moss, D.S., Thornton, J.M. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26: 283–291.
Laskowski, R.A., Watson, J.D., Thornton, J.M. 2005. ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res. 33: 89–93.
Laurie, A. and Jackson, R. 2005. Q-SiteFinder: an energy-based method for the prediction of proteinligand binding sites. Bioinformatics 21: 1908–1916.
Lüthy, R., Bowie, J.U., Eisenberg, D. 1992. Assessment of protein models with three-dimensional profiles. Nature 356: 83–85.
Lippi, M., Passerini, A., Punta, M., Rost B., Frasconi, P. 2008. MetalDetector: a web server for predicting metal-binding sites and disulfide bridges in proteins from sequence. Bioinformatics 24(18): 2094–2095.
Martí-Renom, M.A., Stuart, A.C., Fiser, A., Sánchez, R., Melo, F., Sali, A. 2000. Comparative protein structure modeling of genes and genomes. Annu. Rev. Biophys. Biomol. Struct. 29: 291–325
Matthias Heinig, Dmitrij, Frishman. 2004 STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32: 500–502.
Moller, I.M. 2001. Annu. Rev. Plant. Physiol. Plant. Mol. Biol. 52: 561.
Michener, C.D., Sokal, R.R. 1957. A quantitative approach to a problem of classification. Evolution. 11: 490–499.
Ouyang, S., Zhu W., Hamilton, J., Lin H., Campbell, M., Childs, K., Thibaud-Nissen, F., Malek, R. L., Lee, Y., Zheng, L., Orvis, J., Haas, B., Wortman, J., and Buell, C. R. 2007. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Research. 35: 883–887.
Sakamoto, A., Nosaka, Y. and Tanaka, K. 1993. Cloning and Sequencing Analysis of a Complementary DNA for Manganese-Superoxide Dismutase from Rice (Oryza sativa). Plant Physiol. 103: 1477–1478.
Sali, A., Blundell, T.L. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234: 779–815.
Tamura, K., Dudley, J., Nei, M., Kumar, S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–9.
Thompson, J.D., Higgins, D.G., Gibson, T.J. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 22(22): 4673–80.
Trinh, C.H., Hunter, T., Stewart, E.E., Phillips, S.E., Hunter, G.J. 2008. Purification, crystallization and Xray structures of the two manganese superoxide dismutases from Caenorhabditis elegans. Acta Crystallogr., Sect. F 64: 1110–1114.
Wiberg, K.B. 1965. A scheme for strain energy minimization. J. Am. Chem. Soc. 87: 1070–1078.
Wiederstein, M., Sippl, M.J. 2007. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucl. Acids. Res. 35: 407–410.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tripathi, V., Tripathi, P. Molecular phylogenetics and comparative modeling of MnSOD, an enzyme involved during environmental stress conditions in Oryza sativa . Interdiscip Sci Comput Life Sci 6, 251–258 (2014). https://doi.org/10.1007/s12539-011-0050-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-011-0050-4