Abstract
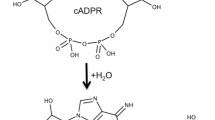
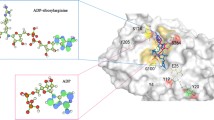
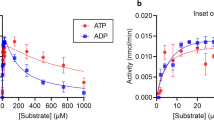
A series of the complexes of human CD38’s wild type, E226 and E146 mutants as well have been simulated. The biosoftwares well simulate the penetration of nicotinamide-adenine-dinucleotide (NAD) into the active site. The nicotinamide end of NAD penetrates deep into the active site consistent with cleavage of the nicotinamide-glycosidic bond which is the first step of catalysis creating a Michaelis complex regarded as the intermediate product of NAD cyclase and hydrolysis reaction. The breaking down hydrogen bond between 2′-3′ OH ribosyl and the residues replaced Glu226 makes NAD to be less constrained in active site and nicotinamide (NA) becomes more difficult to be cleaved and eliminates the mutant catalytic activities. The large majority of the substrate NAD is hydrolyzed to ADPR while the conversion of NAD to cADPR is not the dominant reaction catalyzed by wild-type human CD38. The more strongly kept ribosyl group by hydrogen bonds the more NADase and the less cyclase activity. Breaking hydrogen bonds of ribosyl 2′- and 3′-OH by mutation will loosen it to promote the cyclase. The cyclic adenosine diphosphate-ribose (cADPR) could also penetrate deeply into active site to make some hydrogen bonds with Glu146 and Glu226; however, its docking poses are affected by a residue located at the entrance of the catalytic pocket (Lys129). These results are in good agreement with the previous crystallographic analysis and the experiments quantified the catalytic activities of human CD38 and its mutants.
Similar content being viewed by others
References
Böhm, H.J., Klebe, G. 1996. What can we learn from molecular recognition in protein-ligand complexes for the design of new drugs? Angew Chem Int Ed Engl 35, 2588–2614.
Brooks, B.R., Bruccoleri, R.E., Olafson, B.D., States, D.J., Swaminathan, S., Kaplus, M. 1983. CHARMM: A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem 4, 187–217.
Cavalli, A., Carloni, P., Recanatini M. 2006. Target-related applications of first principles quantum chemical methods in drug design. Chemical Reviews 106, 3497–3519.
Graeff, R., Walseth, T.F., Fryxell, K., Branton, W.D., Lee, H.C. 1994. Enzymatic synthesis and characterizations of cyclic GDP-ribose: A procedure for distinguishing enzymes with ADP-ribosyl cyclase activity. J Biol Chem 269, 30260–30267.
Graeff, R.M., Munshi, C., Aarhus, R., Johns, M., Lee, H.C. 2001. A single residue at the active site of CD38 determines its NAD cyclizing and hydrolyzing activities. J Biol Chem 276, 12169–12173.
Guex, N., Peitsch, M.C. 1997. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis 18, 2714–2723.
Howard, M., Grimaldi, J.C., Bazan, J.F., Lund, F.E., Santos-Argumendo, L., Parkhouse, R.M.E., Walseth, T.F., Lee, H.C. 1993. Formation and hydroysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science 262, 1056–1059.
HYPERCHEM 8.0 Evaluation. 2007. Hypercube, Inc.
Jorgensen, W.L., Tirado-Rives, J. 1988. The OPLS potential functions for proteins. Energy minimizations for crystals of cyclic peptides and Crambin. J Am Chem Soc 110, 1657–1666.
Jorgensen, W.L. 2004. The many roles of computation in drug discovery. Science 303, 1813–1818.
Khoo, K.M., Chang, C.F. 1999. Characterization and localization of CD38 in the vertebrate eye. Brain Res 821, 17–25.
Lee, H.C. 2006. Structure and enzymatic functions of human CD38. Mol Med 12, 317–323.
Liu, Q., Kriksunov, I.A., Graeff, R., Munshi, C., Lee, H.C., Hao, Q. 2005. Crystal structure of human CD38 extracellular domain. Structure 13, 1331–1339.
Liu, Q., Kriksunov, I.A., Graeff, R., Munshi, C., Lee, H.C., Hao, Q. 2006. Structural basis for the mechanistic understanding of human CD38-controlled multiple catalysis. J Biol Chem 281, 32861–32869.
Liu, Q., Kriksunov, I.A., Graeff, R., Lee, H.C., Hao, Q. 2007. Structural basis for formation and hydrolysis of the Calcium messenger cyclic ADP-ribose by human CD38. J Biol Chem 282, 5853–5861.
Malavasi, F., Funaro, A., Roggero, S., Horenstein, A., Calosso, L., Mehta, K. 1994. Human CD38: A glycoprotein in search of a function. Immunol Today 15, 95–97.
Mousseau, N., Derreumaux, P., Barkema, G.T., Malek, R. 2001. Sampling activated mechanisms in proteins with the activation-relaxation technique. J Mol Graphics Modell 19, 78–86.
Munshi, C., Aarhus, R., Graeff, R., Walseth, T.F., Levitt, D., Lee, H.C. 2000. Identification of the enzymatic active site of CD38 by site-directed mutagenesis. J Biol Chem 275, 21566–21571.
Parthiban, V., Gromiha, M.M., Schomburg, D. 2006. CUPSAT: Prediction of protein stability upon point mutations. Nucleic Acids Research 34, W239–W242.
QUANTUM 3.3 Docking/Library Screening Software Quantum Pharmaceuticals 2007.
Sauve, A.A., Deng, H.T., Angeletti, R.H., Schramm, V.L. 2000. A covalent intermediate in CD38 is responsible for ADP-ribosylation and cyclization reactions. J Am Chem Soc 122, 7855–7859.
Schrodinger, L.L.C. 2000. GLIDE, Portland, OR.
Sherwood, P. 2000. Hybrid quantum mechanics/molecular mechanics approaches. In: Grotendorst, J. (Ed) NIC Series Volume 1, Juelich 257–277.
Tohgo, A., Munakata, H., Takasawa, S., Nata, K., Akiyama, T., Hayashi, N., Okamoto, H. 1997. Lysine 129 of CD38 (ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase) participates in the binding of ATP to inhibit the cyclic ADP-ribose hydrolase. J Biol Chem 272, 3879–3882.
Vriend, G. 1990. WHAT IF: A molecular modeling and drug design program. J Mol Graph 8, 52–56.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nguyen, M.H., Dang, V.U. & Luu, B.V. Computational characterization for catalytic activities of human CD38’s wild type, E226 and E146 mutants. Interdiscip Sci Comput Life Sci 2, 193–204 (2010). https://doi.org/10.1007/s12539-010-0091-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-010-0091-0