Abstract
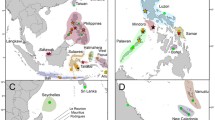
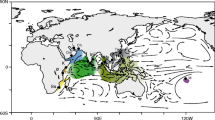
We found that Hippocampus mohnikei, the most common seahorse in East Asia, includes three clades with 2.8%–7.3% pairwise differences in their DNA cytochrome b (mtDNA cyt b) sequences, and all the clades are detected in the eastern Korea Strait (only in Geoje), Korea. Clades A and B have different demographic and divergence features via NJ-tree, TCS, mismatch, and BSP analysis from mtDNA cyt b, and the hybridization among the three clades was detected in nuclear DNA (the 1st intron RP1 of ribosomal protein S7 gene, ncDNA S7), implying a possibility of historic secondary contact or ancestral polymorphism. We also found the 1.4%-divergence-per-million-years molecular clock in mtDNA cyt b is appropriate for H. mohnikei through mismatch and BSP analysis comparison. Disconnection of each clade among the Yellow Sea, Korea Strait, and Japanese waters are related to different water masses, therefore there may exist an incomplete biogeographic barriers among them via discordant results between mtDNA and ncDNA. Therefore, a new analysis (such as microsatellite DNA or RAD-seq) must be considered to determine whether H. mohnikei constitutes a widely distributed panmictic population or several reproductively isolated populations.






Similar content being viewed by others
References
Aylesworth L, Lawson JM, Laksanawimol P, Ferber P, Loh TL (2016) New records of the Japanese seahorse Hippocampus mohnikei in Southeast Asia lead to updates in range, habitat and threats. J Fish Biol 88(4):1620–1630
Bae SE, Kim JK, Kim JH (2016) Evidence of incomplete lineage sorting or restricted secondary contact in Lateolabrax japonicus Complex (Actinopterygii: Moronidae) based on morphological and molecular traits. Biochem Syst Ecol 66:98–108
Bickford D, Lohman DJ, Sodhi NS, Ng PKL, Meier R, Winker K, Ingram KK, Das I (2007) Cryptic species as a window on diversity and conservation. Trend Ecol Evol 22:148–155
Boehm JT, Woodall L, Teske PR, Lourie SA, Baldwin C, Waldman J, Hickerson M (2013) Marine dispersal and barriers drive Atlantic seahorse diversification. J Biogeogr 40(10):1839–1849
Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ (2014) BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10(4):e1003537
Buckley TR, Marske K, Attanayake D (2010) Phylogeography and ecological niche modelling of the New Zealand stick insect Clitarchus hookeri (white) support survival in multiple coastal refugia. J Biogeogr 37(4):682–695
Casey SP, Hall HJ, Stanley HF, Vincent ACJ (2004) The origin and evolution of seahorses (genus Hippocampus): a phylogenetic study using the cytochrome b gene of mitochondrial DNA. Mol Phylogenet Evol 30(2):261–272
Chakraborty A, Iwatsuki Y (2006) Genetic variation at the mitochondrial 16S rRNA gene among Trichiurus lepturus (Teleostei: Trichiuridae) from various localities: preliminary evidence of a new species from west coast of Africa. Hydrobiologia 563(1):501–513
Choi YU, Rho S, Jung MM, Lee YD, Noh GA (2006) Parturition and early growth of crowned seahorse, Hippocampus coronatus in Korea. Korean Journal of Aquaculture 19(2):109–118 (In Korean)
Choo CK, Liew HC (2006) Morphological development and allometric growth patterns in the juvenile seahorse Hippocampus kuda Bleeker. J Fish Biol 69(2):426–445
Chow S, Hazama K (1998) Universal PCR primers for S7 ribosomal protein gene introns in fish. Mol Ecol 7(9):1255–1256
Clement M, Snell Q, Walke P, Posada D, Crandall K (2002) TCS: estimating gene genealogies. Proc 16th Int Parallel Distrib Process Symp 2:184–190
Curtis JMR, Vincent ACJ (2006) Life history of an unusual marine fish: survival, growth and movement patterns of Hippocampus guttulatus Cuvier 1829. J Fish Biol 68:707–733
Drummond AJ, Rambaut A, Shapiro B, Pybus OG (2005) Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol 22(5):1185–1192
Excoffier L, Lischer HE (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and windows. Mol Ecol Resour 10(3):564–567
Foster SJ (2016) Seahorses (Hippocampus spp.) and the CITES review of significant trade. Fish Cent Res Rep 24(4):48
Froese R, Pauly D (2016) FishBase version (01/2016). World Wide Web electronic publication. http://www.fishbase.org/. Accessed 27 June 2016
Fu YX (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147(2):915–925
Grant WS, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. J Hered 89(5):415–426
Grant WS, Liu M, Gao T, Yanagimoto T (2012) Limits of Bayesian skyline plot analysis of mtDNA sequences to infer historical demographies in Pacific herring (and other species). Mol Phylogenet Evol 65(1):203–212
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucl Acid S 41:95–98
Han SY, Lee JM, Kim JK (2014) New record of the short-tailed pipefish, Microphis brachyurus brachyurus (Teleostei: Syngnathidae), with a key to the species of the family Syngnathidae from Korea. Ocean Science Journal 49(4):419–424
He L, Mukai T, Chu KH, Ma Q, Zhang J (2015) Biogeographical role of the Kuroshio current in the amphibious mudskipper Periophthalmus modestus indicated by mitochondrial DNA data. Sci Rep 5:15645
Ho SY, Shapiro B (2011) Skyline-plot methods for estimating demographic history from nucleotide sequences. Mol Ecol Resour 11(3):423–434
Hyde JR, Underkoffler KE, Sundberg MA (2014) DNA barcoding provides support for a cryptic species complex within the globally distributed and fishery important opah (Lampris guttatus). Mol Ecol Resour 14(6):1239–1247
Jeong HD, Kwoun CH, Kim SW, Cho KD (2009) Fluctuation of tidal front and expansion of cold water region in the southwestern sea of Korea. Journal of the Korean Society of Marine Environment & Safety 15(4):289–296 (In Korean)
Kafanov AI, Volvenko IV, Fedorov VV, Pitruk DL (2000) Ichthyofaunistic biogeography of the Japan (East) Sea. J Biogeogr 27:915–933
Kafanov AI, Volvenko IV, Pitruk DL (2001) Ichthyofaunistic biogeography of the East Sea: comparison between benthic and pelagic zonalities. Ocean and Polar Research 23(1):35–49
Kai Y, Park KD, Nakabo T (2012) The incomplete history of mitochondrial lineages between two rockfishes, Sebastes longispinis and Sebastes hubbsi (Scorpaeniformes: Scorpaenidae). J Fish Biol 81(3):954–965
Kanou K, Kohno H (2001) Early life history of a seahorse, Hippocampus mohnikei, in Tokyo Bay, Japan. Ichthyol Res 48(4):361–368
Keigwin LD (1978) Pliocene closing of the isthmus of Panama, based on biostratigraphic evidence from nearby Pacific Ocean and Caribbean Sea cores. Geology 6(10):630–634
Kim JK, Choi OI, Chang DS, Kim JI (2002) Fluctuation of bag-net catches off Wando, Korea and the effect of sea water temperature. Kor J Fish Aquat Sci 35(5):497–503 (In Korean)
Kim IS, Choi Y, Lee CL, Lee YJ, Kim BJ, Kim JH (2005) Illustrated book of Korean fishes. Kyohaksa, Seoul (In Korean)
Kim JK (2009) Diversity and conservation of Korean marine fishes. Korean J Ichthyol 21(Supplement):52–62
Kim JK, Bae SE, Lee SJ, Yoon MG (2017) New insight into hybridization and unidirectional introgression between Ammodytes japonicus and Ammdytes heian (Pisces, Ammodytidae). PLoS One 12(6):e0178001
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16(2):111–120
Koh JR, Kim DH, Yun SJ, Oh TY (2004) Early life history and rearing of the yellow seahorse Hippocampus kuda (Teleostei: Syngnathidae). Korean J Ichthyol 16(1):1–8 (In Korean)
Li Y, Qian ZJ, Kim SK (2008) Cathepsin B inhibitory activities of three new phthalate derivatives isolated from seahorse, Hippocampus Kuda Bleeler. Bioorg Med Chem Lett 18(23):6130–6134
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25(11):1451–1452
Liu JX, Gao TX, Wu SF, Zhang YP (2007) Pleistocene isolation in the Northwestern Pacific marginal seas and limited dispersal in a marine fish, Chelon haematocheilus (Temminck & Schlegel, 1845). Mol Ecol 16(2):275–288
Lorenzo-Carballa MO, Hadrys H, Cordero-Rivera A, Andrés JA (2012) Population genetic structure of sexual and parthenogenetic damselflies inferred from mitochondrial and nuclear markers. Heredity 108(4):386–395
Lourie SA, Vincent ACJ, Hall HJ (1999) Seahorses: an identification guide to the world’s species and their conservation. Project Seahorse, London
Lourie SA, Foster SJ, Cooper EWT, Vincent ACJ (2004) A guide to the identification of seahorses. Project Seahorse and TRAFFIC North America, University of British Columbia and World Wildlife Fund, Washington DC
Lourie SA, Vincent ACJ (2004) A marine fish follows Wallace’s line: the phylogeography of the three-spot seahorse (Hippocampus trimaculatus, Syngnathidae, Teleostei) in Southeast Asia. J Biogeogr 31(12):1975–1985
Lourie SA, Green DM, Vincent ACJ (2005) Dispersal, habitat differences, and comparative phylogeography of southeast Asian seahorses (Syngnathidae: Hippocampus). Mol Ecol 14(4):1073–1094
Lourie SA, Pollom RA, Foster SJ (2016) A global revision of the seahorses Hippocampus Rafinesque 1810 (Actinopterygii: Syngnathiformes): taxonomy and biogeography with recommendations for further research. Zootaxa 4146(1):1–66
Luzzatto DC, Estalles ML, Díaz de Astarloa JM (2013) Rafting seahorses: the presence of juvenile Hippocampus patagonicus in floating debris. J Fish Biol 83(3):677–681
Myoung SH, Kim JK (2014) Genetic diversity and population structure of the gizzard shad, Konosirus punctatus (Clupeidae, Pisces), in Korean waters based on mitochondrial DNA control region sequences. Genes & Genomics 36(5):591–598
Nakabo T (2009) Zoogeography and systematics of shallow water marine East Asian fishes. Korean J Ichthyol 21(s):38–43
Nyakaana S, Tumusiime C, Oguge N, Siegismund HR, Arctander P, Muwanika V (2008) Mitochondrial DNA diversity and population structure of a forest-dependent rodent, Praomys taitae (Rodentia: Muridae) Heller 1911, in the fragmented forest patches of Taita Hills, Kenya. S Afr J Sci 104(11–12):499–504
Rambaut A, Suchard MA, Xie D, Drummond AJ (2014) Trace:v1.6. Available from http://beast.bio.ed.ac.uk/Tracer
Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19(12):2092–2100
Rebstock GA, Kang YS (2003) A comparison of three marine ecosystems surrounding the Korean peninsula: responses to climate change. Prog Oceanogr 59(4):357–379
Rogers AR, Harpending H (1992) Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol 9(3):552–569
Sakaguchi SO, Ueda H, Ohtsuka S, Soh HY, Yoon YH (2011) Zoogeography of planktonic brackish-water calanoid copepods in western Japan with comparison with neighboring Korean fauna. Plankton and Benthos Research 6(1):18–25
Schneider S, Excoffier L (1999) Estimation of past demographic parameters from the distribution of pairwise differences when the mutation rates vary among sites: application to human mitochondrial DNA. Genetics 152(3):1079–1089
Senou H (2013) Syngnathidae. In: Nakabo T (ed) Fishes of Japan with pictorial keys to the species, 3rd edn. Tokai University Press, Kanagawa, pp 615–635 (In Japanese)
Sonnenberg R, Nolte AW, Tautz D (2007) An evaluation of LSU rDNA D1-D2 sequences for their use in species identification. Front Zool 4(1):6
Sousa-Santos C, Robalo JI, Collares-Pereira MJ, Almada VC (2005) Heterozygous indels as useful tools in the reconstruction of DNA sequences and in the assessment of ploidy level and genomic constitution of hybrid organisms. DNA Seq 16(6):462–467
Spalding MD, Fox HE, Allen GR, Davidson N, Ferdaña ZA, Finlayson M, Halpern BS, Jorge MA, Lombana A, Lourie SA, Martin KD, McManus E, Molnar J, Recchia CA, Robertson J (2007) Marine ecoregions of the world: a bioregionalization of coastal and shelf areas. Bioscience 57(7):573–583
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Teske PR, Cherry MI, Matthee CA (2004) The evolutionary history of seahorses (Syngnathidae: Hippocampus): molecular data suggest a West Pacific origin and two invasions of the Atlantic Ocean. Mol Phylogenet Evol 30(2):273–286
Teske PR, Hamilton H, Palsboll PJ, Choo CK, Gabr H, Lourie SA, Santos M, Sreepada A, Cherry MI, Matthee CA (2005) Molecular evidence for long-distance colonization in an indo-Pacific seahorse lineage. Mar Ecol Prog Ser 286:249–260
Thornhill DJ (2012) Ecological impacts and practices of the coral reef wildlife trade. Defenders of Wildlife, Washington DC
Vandendriessche S, Messiaen M, Vincx M, Degraer S (2005) Juvenile Hippocampus guttulatus from a neuston tow at the French-Belgian border. Belg J Zool 135(1):101–102
Vincent ACJ (1996) The international trade in seahorses. Traffic International, Cambridge
Wan S, Li A, Clift PD, Stuut JBW (2007) Development of the east Asian monsoon: mineralogical and sedimentologic records in the northern South China Sea since 20 ma. Palaeogeogr Palaeoclimatol Palaeoecol 254(3):561–582
Wang P, Sun X (1994) Last glacial maximum in China: comparison between land and sea. Catena 23(3):341–353
Yan Q, Shi X (2014) Petrologic perspectives on tectonic evolution of a nascent basin (Okinawa Trough) behind Ryukyu Arc: a review. Acta Oceanol Sin 33(4):1–12
Yang YJ, Kim SK, Park SJ (2012) An anti-inflammatory peptide isolated from seahorse Hippocampus kuda bleeler inhibits the invasive potential of MG-63 Osteosarcoma cells. Fisheries and Aquatic Sciences 15(1):29–36
Yoo JT, Kim JK, Choi MS (2014) Change of structure community of fish collected by a gape net with wings after 12 years in the coast of Wando Island, Korea. Kor J Fish Aquat Sci 47:659–666 (In Korean)
Zhang Y, Pham NK, Zhang H, Lin J, Lin Q (2014) Genetic variations in two seahorse species (Hippocampus mohnikei and Hippocampus trimaculatus): evidence for middle Pleistocene population expansion. PLoS One 9(8):e105494
Acknowledgements
This research was supported by a grant from the Marine Fish Resources Bank of Korea (MFRBK) and by a grant from the National Institute of Fisheries Science, Korea (NIFS, R2017032). We would like to deeply thank M. S. Choi (Southwest Sea Fisheries Research Institute, Korea), J. H. Park (NIFS, Korea), W. G. Park and J. Y. Bae (Zooplanktology lab. of Pukyong National University, Korea), S. Rho, G. E. Noh, and S. O. Shin (Haecheonma, Korea), K. S. Han (Dongwon Institute of Science and Technology, Korea), H. Sugawara, H. D. Mun, G. H. Mun, Y. G. Park, M. S. Kim, and J. H. Lee for help in our sampling.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by H. Stuckas
Rights and permissions
About this article
Cite this article
Han, SY., Kim, JK., Tashiro, F. et al. Relative importance of ocean currents and fronts in population structures of marine fish: a lesson from the cryptic lineages of the Hippocampus mohnikei complex. Mar Biodiv 49, 263–275 (2019). https://doi.org/10.1007/s12526-017-0792-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12526-017-0792-2