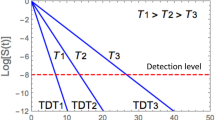
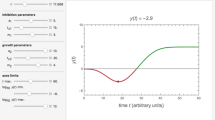
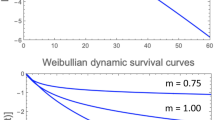
Abstract
Regardless of the targeted microbe type, a thermal or nonthermal food preservation or disinfection method’s efficacy is primarily assessed based on its kinetics. Yet, there is growing realization that inactivation kinetics and the individual microbes’ spectrum of vulnerabilities or resistances to a lethal agent are two sides of the same coin. This creates the possibility to convert traditional survival data plotted on linear or semilogarithmic coordinates to temporal distributions of the individual microbes’ deactivation, or vice versa. Such conversions are demonstrated with simulated microbial survival patterns generated with different kinds of survival models: the two-parameter Weibull distribution of which the single-parameter loglinear model is a special case, the normal, lognormal, and Fermi distribution functions, which imply that complete microbial inactivation is theoretically impossible, the three-parameter Gompertz survival model which allows for definite residual survival, and the three-parameter version of the beta distribution function, allowing for a definite thermal death time beyond which no survivors will ever be found. Also provided are simulated examples of the survival patterns of mixed microbial populations, and they all demonstrate that the common shapes of microbial survival curves do not contain enough information to infer whether the targeted microbial population is genetically or physiologically uniform or a mixture of subpopulations. The presented analysis lends support to the notion that any proposed microbial survival kinetic model’s validity should be tested by its ability to predict survival patterns not used in its formulation and not by statistical fit criteria.













Similar content being viewed by others
Availability of Data and Materials
Not applicable.
References
Van Boekel MAJS (2002) On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Intnt J Food Microbiol 74:139–159
Van Boekel MAJS (2008) Kinetic modeling of food quality: a critical review. Comprehensive Rev Food Sci Food Safety 7:144–158
Peleg M, Cole MB (1998) Reinterpretation of microbial survival curves. Crit Rev Food Sci Nutr 38:353–380
Peleg M (2003) Microbial survival curves: interpretation, mathematical modeling, and utilization. Comment Theor Biol 8:357–387
Mafart P, Couvert O, Gaillard S, Leguerinel I (2002) On calculating sterility in thermal preservation methods: application of the Weibull frequency distribution model. Internl J Food Microbiol 72:107–113
Peleg M (1996) Evaluation of the Fermi equation as a model of dose-response curves. Appl Microbiol Biotechnol 46:303–306
Peleg M (2017) Modeling microbial inactivation by pulsed electric fields. In: Miklavcic D (ed) Handbook of electroporation. Springer, pp 1269–1286
Koyama K, Hiroki H, Kawamura S, Koseki S (2019) Calculating stochastic inactivation of individual cells in a bacterial population using variability in individual cell inactivation time and initial cell number. J Theor Biol 469:172–179
Zwietering MH, Garre A, den Besten HMW (2021) Incorporating variability in the design of heat treatment: a stochastic approach and kinetic approach. Food Res Intnl 139:109973
Garre A, den Besten HMW, Hernandez PS, Zwietering MH (2020) Not just variability and uncertainty: the relevance of chance for the survival of microbial cells to stress. Trends Food Sci Technol 118:799–807
Aspridou Z, Koutsoumanis K (2020) Variability in microbial inactivation: from deterministic Bigelow model to probability distribution of single cell inactivation times. Food Res Intnl 137:109579
Peleg M (2006) Advanced quantitative microbiology for food and biosystems: models for predicting growth and inactivation. CRC Press, Boca Raton FL
Aragao GMF, Corradini MG, Normand MD, Peleg M (2007) Evaluation of the Weibull and log-normal distribution functions as survival models of Escherichia coli under isothermal and non-isothermal conditions. Intnl J Food 19:243–257
Peleg M (2023) Selected challenges to modeling the kinetics of microbial inactivation and chemical reactions during food preservation. Curr Opin Food Sci 51:101029
Arnoldini M, Vizcarra IA, Peña-Miller R, Stocker N, Diard M (2014) Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol 12(8):e1001928. https://doi.org/10.1371/journal.pbio.1001928
Tuomanen E, Cozens R, Tosch W, Zak O, Tomasz A (1986) The rate of killing of Escherichia coli by ß-lactam antibiotics is strictly proportional to the rate of bacterial growth. J Gen Microbiol 132:1297–1304
Horowitz J, Normand MD, Corradini MG, Peleg M (2010) A probabilistic model of growth, division, and mortality of microbial cells. Appl Environ Microbiol 76:230–242
Corradini MG, Normand MD, Peleg M (2010) Stochastic and deterministic model of microbial heat inactivation. J Food Sci 75:R59–R70
Stone G, Chapman B, Lowel D (2009) Development of a log-quadratic model to describe microbial inactivation, illustrated by thermal inactivation of Clostridium botulinum. Appl Environ Microbiol 75:6998–7005
Acknowledgements
The author expresses his deep gratitude to Mark D. Normand for his valuable help in programming.
Author information
Authors and Affiliations
Contributions
The author wrote the paper and produced the figures.
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Competing Interests
The author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peleg, M. Microbial Inactivation Kinetics Models, Survival Curves Shapes, and the Temporal Distributions of the Individual Germs Deactivation. Food Eng Rev (2024). https://doi.org/10.1007/s12393-024-09367-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12393-024-09367-5