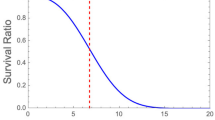
Abstract
During microbial inactivation by a lethal agent (ignoring damage/injury, recovery from damage/injury, etc.), an individual spore or cell can be either viable/alive and countable, or already inactivated/dead and uncountable. But since we only count the survivors’ total number at successive times, the fates of the individual microbes remain unknown. This makes the process probabilistic and creates the need for a stochastic model to describe it. The familiar continuous deterministic models such as the loglinear or Weibullian, which are only applicable to large populations, can be viewed as the mathematical limits of underlying discrete stochastic models. When the targeted microbial population is initially small, its survival curve is inherently irregular and irreproducible. But when the targeted population is large, its survival curve is initially smooth and reproducible but inevitably becomes irregular and irreproducible as the number of survivors diminishes. Perhaps the most important difference between the deterministic loglinear model, or the Weibullian with a shape factor > 1, and their fully stochastic versions is that the latter predict complete elimination of the targeted microbe in a realistic finite time. A stochastic survival model is derived from the individual microbes’ Markov chains (or trees) and the character of the underlying survival probability rate’s time-dependence. Various types of such dependencies are presented, and their different manifestations in the stochastic survival curves shapes are demonstrated. Also discussed are ways to extract (estimate) the stochastic model’s parameters from the regular and reproducible part of static experimental survival data, which in some cases requires unconventional regression techniques.










Similar content being viewed by others
References
Buzrul S (2022) The Weibull model for microbial inactivation. Food Eng Rev 14:45–61
Mafart P, Couvert O, Gaillard S, Leguerinel I (2002) On calculating sterility in thermal preservation methods: application of the Weibull frequency distribution model. Intnl J Food Microbiol 72:107–113
Peleg M (2006) Advanced quantitative microbiology for foods and biosystems: models for predicting growth and inactivation. Taylor and Francis, Boca Raton, FL
Peleg M, Cole MB (1998) Reinterpretation of microbial survival curves. Crit Rev Food Sci Nut 38:353–380
van Boekel MAJS (2002) On the use of the Weibull model to describe thermal inactivation of microbial vegetative cells. Intnl J Food Res 74:139–215
Horowitz J, Normand MD, Corradini MG, Peleg M (2010) A probabilistic model of growth, division, and mortality of microbial cells. Appl Environ Microbiol 76:230–242
Corradini MG, Normand MD, Peleg M (2010) A stochastic and deterministic model of microbial heat inactivation. J Food Sci 75:R59–R70
Corradini MG, Normand MD, Eisenberg M, Peleg M (2010) Evaluation of a stochastic inactivation model for heat-activated spores of Bacillus spp. Appl Environ Microbiol 76:4402–4412
Koutsoumanis KP, Asprido Z (2017) Individual cell heterogeneity in predictive food microbiology: challenges in predicting a “noisy” world. Intnl J Food Micorbiol 240:3–10
Garre A, den Besten HMW, Fernandez P, Zwietering MH (2021) Not just variability and uncertainty; the relevance of chance for the survival of microbial cells to stress. Trends Food Sci Technol 112:799–807
Koseki S, Koyama K, Abe H (2021) Recent advances in predictive microbiology: theory and application of conversion from population dynamics to individual cell heterogeneity during inactivation process. Curr Opinion Food Sci 39:60–67
Peleg M (2021) The thermal death time concept and its implications revisited. Food Eng Rev 13:291–304
Peleg M (2020) Endpoints method for predicting microbial inactivation, nutrients degradation and quality loss at high and ultra-high temperatures. In: Demirci A, Feng H, Krishnamurthy K (eds) Food Safety Engineering. Springer, pp 421–446
Peleg M, Normand MD, Corradini MG, van Asselt AJ, de Jong P, ter Steeg PF (2008) Estimating the heat resistance parameters of bacterial spores from their survival ratios at the end of UHT and other heat treatments. Crit Rev Food Sci Nut 48:634–648
Peleg M, Penchina CM (2000) Modeling microbial survival during exposure to a lethal agent with varying intensity. Crit Rev Food Sci Nut 40:159–172
Acknowledgements
The author expresses his deep gratitude to Mark D. Normand who has programmed all the mathematical software used or cited in this review.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The author declares no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peleg, M. Fully Probabilistic Microbial Inactivation Models: the Markov Chain Reconstruction from Experimental Survival Ratios. Food Eng Rev 15, 1–14 (2023). https://doi.org/10.1007/s12393-022-09325-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12393-022-09325-z