Abstract
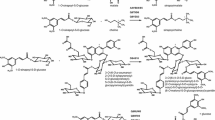
O-Methyltransferases (OMTs) transfer a methyl group from S-adenosylmethionine to a hydroxyl group of an acceptor. One group of OMTs is the caffeoyl-CoA O-methyltransferase type, which is involved in the biosynthesis of monolignol. In this study, OsOMT26 was cloned from Oryza sativa and the recombinant OsOMT26 protein was characterized. OsOMT26 used not only caffeoyl-CoA as a substrate but also different flavonoids such as luteolin and tricetin. However, when caffeoyl-CoA was used as the substrate, the reactivity of OsOMT26 was approximately 6.6-fold better than when either luteolin or tricetin was used. This result demonstrated that OsOMT26 displayed the typical properties characteristic of CCoAOMT. Molecular modeling followed by site-directed mutagenesis was employed to examine why caffeic acid or caffeoyl-CoA was a better substrate than tricetin. One amino acid, Asp210, turned out to be critical for substrate binding, and site-directed mutagenesis of Asp to Glu improved the enzyme’s reactivity toward tricetin.



Similar content being viewed by others
Abbreviations
- CCoAOMT:
-
Caffeoyl-CoA OMT
- COMTs:
-
Caffeic acid OMTs
- 4-CL:
-
4-Coumaroyl-CoA ligase
- OMT:
-
O-methyltransferase
- SAM:
-
S-adenosylmethionine
References
Bheemanaik S, Reddy YV, Rao DN (2006) Structure, function and mechanism of exocyclic DNA methyltransferases. Biochem J 399:177–190
Bügl H, Fauman EB, Staker BL, Zheng F, Kushner SR, Saper MA, Bardwell JC, Jakob U (2002) RNA methylation under heat shock control. Mol Cell 6:349–360
Cho J-H, Park Y, Ahn J-H, Lim Y, Rhee S (2008) Structural and functional insights of O-methyltransferase from Bacillus cereus. J Mol Biol 382:987–997
Ferrer F-L, Zubieta C, Dixon RA, Noel JP (2005) Crystal structures of alfalfa caffeoyl coenzyme A 3-O-methyltransferase. Plant Physiol 137:1009–1017
Ibrahim RK, De Luca V, Khouri HE, Latchinian L, Brisson L, Charest PM (1987) Enzymology and compartmentation of polymethylated flavonol glucosides in Chrysosplenium americanum. Phytochemistry 26:1237–1245
Joe EJ, Kim B-G, Ahn B-C, Chong Y, Ahn J-H (2010) Engineering of flavonoid O-methyltransferase for a novel regioselectivity. Mol Cell 30:137–141
Joshi CP, Chiang VL (1998) Conserved sequence motifs in plant S-adenosyl-l-methionine-dependent methyltransferases. Plant Mol Biol 37:663–674
Kim B-G, Sung SH, Chong Y, Lim Y, Ahn J-H (2010) Plant flavonoid O-methyltransferases: substrate specificity and application. J Plant Biol 53:321–329
Kim BG, Lee Y, Hur H-G, Lim Y, Ahn J-H (2006a) Flavonoid 3′-O-methyltransferase from rice: cDNA cloning, characterization and functional expression. Phytochemistry 67:387–394
Kim BG, Kim H, Hur HG, Lim Y, Ahn JH (2006b) Regioselectivity of 7-O-methyltransferase of poplar to flavones. J Biotech 138:155–162
Kim DH, Kim BG, Park SH, Kim NY, Lee YJ, Min SY, Park Y-B, Lee J-B, Kim J-C, Lim Y, Chong Y, Ahn J-H (2009) O-Methyltransferase from soybean uses both caffeoyl-CoA and flavonoids as substrates. J Kor Soc Appl Biol Chem 52:114–120
Kim JH, Cheon YM, Kim BG, Ahn J-H (2008) Analysis of flavonoids and characterization of the OsFNS gene involved in flavone biosynthesis in rice. J Plant Biol 51:97–101
Kopycki JG, Rauh D, Chumanevich AA, Neumann P, Vogt T, Stubbs MT (2008) Biochemical and structural analysis of substrate promiscuity in plant Mg2+-dependent O-methyltransferase. J Mol Biol 378:154–164
Lam KC, Ibrahim RK, Behdad B, Dayanandan S (2007) Structure, function, and evolution of plant O-methyltransferase. Genome 50:1001–1013
Lee YJ, Jeon Y, Lee JS, Kim BG, Lee CH, Ahn J-H (2007) Enzymatic synthesis of phenolic CoAs using 4-coumarate:coenzyme A ligase (4CL) from rice. Bull Kor Chem Soc 28:365–366
Lee YJ, Kim BG, Lim Y, Chenog Y, Ahn J-H (2008) Cation dependent O-methyltransferases from rice. Planta 227:641–647
Rastogi S, Dwivedi UN (2008) Manipulation of lignin in plants with special reference to O-methyltransferase. Plant Sci 174:264–277
Schubert HL, Blumenthal RM, Cheng X (2003) Many paths to methyltransfer: a chronicle of convergence. Trends Biochem Sci 28:329–335
Weng J-K, Banks JA, Chapple C (2008) Parallels in lignin biosynthesis. Comm Integr Biol 1:20–22
Whetten R, Sederoff R (1995) Lignin biosynthesis. Plant Cell 7:1001–1013
Acknowledgments
This work was supported by a grant from Systems and Synthetic Agro-biotech Center through Next-Generation BioGreen 21 Program (PJ007975), Agenda program (NIAS, 8-21-52), Rural Development Administration, Republic of Korea, and also partially by the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2009-0093824).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Sung, S.H., Kim, BG., Chong, Y. et al. Characterization of Phenylpropanoid O-Methyltransferase from Rice: Molecular Basis for the Different Reactivity Toward Different Substrates. J. Plant Biol. 54, 314–320 (2011). https://doi.org/10.1007/s12374-011-9169-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12374-011-9169-4