Abstract
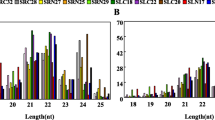
Although sugar beet has a high salt tolerance, the quantity and characteristics of miRNAs and their potential effects on salt stress response remain unclear. The leaves of beet variety "N98122" were used for miRNA sequencing at 0, 2.5, 7.5, and 16h. The results showed that small RNAs of 21 nt had the highest abundance, followed by those of 22 and 23 nt. In total, 654 differential miRNAs, comprising 24 known and 630 novel miRNAs, were determined from seedlings treated with NaCl for 0, 2.5, 7.5, and 16h. Of the 24 known miRNAs, 17 exhibited downregulated expression, including miR159, miR166, miR1873, miR319, miR3626, miR3699, and miR397, and seven miRNAs exhibited upregulated expression, including miR156, miR3630, miR481, miR4995, miR5369, miR536, and miR6173. GRF, HKT, PAGR, SDK, Eps1511, VPS, PAN, 1-Sep, CFDP, DRG, DET, IFRD, PSA, mfsd, and ANTR were determined as the target genes of the differentially expressed miRNAs. The outcomes of this study offer important information about miRNAs that respond to salt conditions, and this knowledge may prove useful for enhancing the tolerance of sugar beet to salt stress.



Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2021) in National Genomics Data Center (Nucleic Acids Res 2022), China National Center for Bioinformation / Beijing Institute of Genomics, Chinese Academy of Sciences (GSA: CRA010166) that are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
References
Bartel, D.P. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297.
Burchett, S.A., P. Flanary, C. Aston, L.X. Jiang, K.H. Young, P. Uetz, S. Fields, and H.G. Dohlman. 2002. Regulation of stress response signaling by the N-terminal dishevelled/EGL-10/Pleckstrin domain of Sst2, a regulator of G protein signaling in saccharomyces cerevisiae. Journal of Biological Chemistry 277: 22156–22167.
Chen, X.B., Z.L. Zhang, D.M. Liu, K. Zhang, A.L. Li, and L. Mao. 2010. SQUAMOSA Promoter-Binding Protein-Like transcription factors: Star players for plant growth and development. Journal of Integrative Plant Biology 52: 946–951.
Dong, B., H.B. Wang, A.P. Song, T. Liu, Y. Chen, W.M. Fang, S.M. Chen, F.D. Chen, Z.Y. Guan, and J.F. Jiang. 2016. miRNAs are involved in determining the improved vigor of autotetrapoid chrysanthemum nankingense. Frontiers in Plant Science, 7.
Fu, C., Y.G. Sun, and G.R. Fu. 2013. Advances of salt tolerance mechanism in hylophyate plants. Biotechnology Bulletin 1: 1–7.
Frazier, T.P., G.L. Sun, C.E. Burklew, and B.H. Zhang. 2011. Salt and drought stresses induce the aberrant expression of microRNA genes in tobacco. Molecular Biotechnology 49: 159–165.
Gu, X., C.M. Ren, B. Liu, J. Li, L. Yang, and J.T. Zhao. 2016. Research on renovation and utilization of saline-alkali land. Heilongjiang Agricultural Sciences 4: 35–38.
Hossain, M.S., A.I. ElSayed, M. Moore, and Karl-Josef. Dietz. 2017. Redox and reactive oxygen species network in acclimation for salinity tolerance in sugar beet. Journal of Experimental Botany 68: 1283–1298.
Hu, Y., J.C. Han, and Y. Zhang. 2015. Summary of saline alkali land improvement techniques. Shanxi Jorunal of Agricultural Sciences 61: 67–71.
Khraiwesh, B., J.K. Zhu, and J.H. Zhu. 2012. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms 1819: 137–148.
Kitazumi, A., Y. Kawahara, T.S. Onda, D.D. Koeyer, and B.G. De Los Reyes. 2015. Implications of miR166 and miR159 induction to the basal response mechanisms of an andigena potato ( Solanum tuberosum subsp. andigena ) to salinity stress, predicted from network models in Arabidopsis. Edited by S. Cloutier. Genome 58: 13–24.
Kong, T., D.S. Zhang, H. Xu, and L.H. Wang. 2014. Microbial ecological characteristics of alkaline-saline lands and its amelioration process: A review. Soils 46: 581–588.
Kuang, L.H., Q.F. Shen, L.Y. Wu, J.H. Yu, L.B. Fu, D.Z. Wu, and G.P. Zhang. 2019. Identification of microRNAs responding to salt stress in barley by high-throughput sequencing and degradome analysis. Environmental and Experimental Botany 160: 59–70.
Leung, A.K.L., and P.A. Sharp. 2010. MicroRNA functions in stress responses. Molecular Cell 40: 205–215.
Li, H.Y., G.Z. Xu, C. Yang, L. Yang, and Z.C. Liang. 2019. Genome-wide identification and expression analysis of HKT transcription factor under salt stress in nine plant species. Ecotoxicology and Environmental Safety 171: 435–442.
Li, M.Y., L.C. Ren, Z. Zou, W. Hu, S.H. Xiao, X.L. Yang, Z.H. Ding, Y. Yan, W.W. Tie, J.H. Yang, and A.P. Guo. 2021. Identification and expression analyses of the special 14–3-3 gene family in papaya and its involvement in fruit development, ripening, and abiotic stress responses. Biochemical Genetics 59: 1599–1616.
Liu, Y.R., D.Y. Li, J.P. Yan, K.X. Wang, H. Luo, and W.J. Zhang. 2019. MiR319 mediated salt tolerance by ethylene. Plant Biotechnology Journal 17: 2370–2383.
Livak, K.J., and T.D. Schmittgen. 2001. Analysis of relative gene expression data using Real-Time Quantitative PCR and the 2−ΔΔCT method. Methods 25: 402–408.
López-Galiano, M.J., I. López-Galiano, A.I. González-Hernández, G. Camañes, B. Vicedo, M.D. Real, and C. Rausell. 2019. Expression of miR159 is altered in tomato plants undergoing drought stress. Plants 8: 201–209.
Lv, X., H. Xu, L. Li, and Y.T. Zhao. 2012. Agricultural sustainable utilization and evaluation of saline-alkali land. Soils 44: 203–207.
Ning, L.H., W.K. Du, H.N. Song, H.B. Shao, W.C. Qi, M.S.A. Sheteiwy, and D.Y. Yu. 2019. Identification of responsive miRNAs involved in combination stresses of phosphate starvation and salt stress in soybean root. Environmental and Experimental Botany 167: 103823.
Rajagopalan, R., H. Vaucheret, J. Trejo, and D.P. Bartel. 2006. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes and Development 20: 3407–3425.
Shan, T., R. Fu, Y. Xie, Q. Chen, Y. Wang, Zh. Li, X. Song, P. Li, and B. Wang. 2020. Regulatory mechanism of maize (Zea mays L.) miR164 in salt stress response. Russian Journal of Genetics 56: 835–842.
Wang, M., Q.L. Wang, and B.H. Zhang. 2013. Response of miRNAs and their targets to salt and drought stresses in cotton (Gossypium hirsutum L.). Gene 530: 26–32.
Wang, Y., T. Zhou, C.P. Yue, J.Y. Huang, and Y.P. Hua. 2022. Identification of miRNAs and their target genes in response to salt stress in Brassica napus. Chinese Journal of Oil Crop Sciences 44: 103–115.
Wei, L.Q., L.F. Yan, and T. Wang. 2011. Deep sequencing on genome-wide scale reveals the unique composition and expression patterns of microRNAs in developing pollen of Oryza sativa. Genome Biology 12: R53.
Wu, G.Q., R.J. Feng, S.J. Li, C.M. Wang, Q. Jiao, and H.L. Liu. 2017. Effects of salt treatments on growth and osmoregulatory substance accumulation in sugar beet (Beta vulgaris). Acta Prataculturae Sinica 26: 169–177.
Xia, J., X.Q. Wang, Y.K. Pierre-François Perroud, R. Quatrano. He, and W.X. Zhang. 2016. Endogenous small-noncoding RNAs and potential functions in desiccation tolerance in physcomitrella patens. Scientific Reports 6: 30118.
Yang, L., C.Q. Ma, L.L. Wang, S.X. Chen, and H.Y. Li. 2012. Salt stress induced proteome and transcriptome changes in sugar beet monosomic addition line M14. Journal of Plant Physiology 169: 839–850.
Zhang, Z.Q., C. Bai, H.Z. Zhang, X.D. Li, Z.J. Fu, S.M. Zhao, and Y.Y. E, H. Zhang, L. Wang and B.Z. Zhang. 2020. Ecaluation and screening of salt tolerance of sugar beet germplasm resources at seedling stage. Chinese Agricultural Science Bulletin 36: 23–28.
Funding
The work was supported by the earmarked fund for CARS (CARS-170104); Inner Mongolia Autonomous Region "the open competition mechanism to select the best candidates" project entitled "Creation of Elite Beet Germplasm and Breeding of Varieties Suitable for Mechanized Operation" (2022JBGS0029); Inner Mongolia key laboratory of sugar beet genetics and germplasm enhancement; Inner Mongolia Natural Science Fund (2023LHMS03031).
Author information
Authors and Affiliations
Contributions
ZZQ contributed to design experiments, stress treatment, analyze data, and write papers; FZJ, CWJ, and WJ involved in data curation and contributed to resources; ZSM, EYY, ZH, ZBZ, and SMY helped to analyze data; ZWZ, DJ, ZZQ, and NZH done partial experiment; LXD and ZHZ involved in argumentation method. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, Z., Fu, Z., Chen, W. et al. Salt Stress Induces Complicated miRNA Responses of Sugar Beet (Beta vulgaris L.). Sugar Tech (2023). https://doi.org/10.1007/s12355-023-01341-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12355-023-01341-5