Abstract
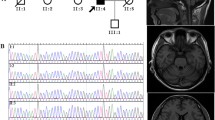
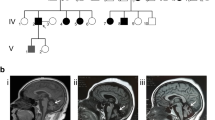
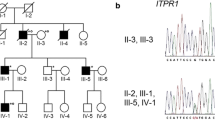
Mutations in STUB1 have been identified to cause autosomal recessive spinocerebellar ataxia type 16 (SCAR16), also named as Gordon Holmes syndrome, which is characterized by cerebellar ataxia, cognitive decline, and hypogonadism. Additionally, several heterozygous mutations in STUB1 have recently been described as a cause of autosomal dominant spinocerebellar ataxia type 48. STUB1 encodes C-terminus of HSC70-interacting protein (CHIP), which functions as an E3 ubiquitin ligase and co-chaperone and has been implicated in several neurodegenerative diseases. In this study, we identified two SCAR16 pedigrees from 512 Taiwanese families with cerebellar ataxia. Two compound heterozygous mutations in STUB1, c.[433A>C];[721C>T] (p.[K145Q];[R241W]) and c.[433A>C];[694T>G] (p.[K145Q];[C232G]), were found in each SCAR16 family by Sanger sequencing, respectively. Among them, STUB1 p.R241W and p.C232G were novel mutations. SCAR16 seems to be an uncommon ataxic syndrome, accounting for 0.4% (2/512) of our cohort with cerebellar ataxia. Clinically, the three patients from the two SCAR16 families presented with cerebellar ataxia alone or in combination with cognitive impairment. The brain MRIs showed a marked cerebellar atrophy of the patients. In conclusion, SCAR16 is an important but often neglected diagnosis of cerebellar ataxia of unknown cause, and the isolated cerebellar ataxia without involvement of other systems cannot be a basis to exclude the possibility of STUB1-related disease.



Similar content being viewed by others
References
Shi Y, Wang J, Li JD, Ren H, Guan W, He M, et al. Identification of CHIP as a novel causative gene for autosomal recessive cerebellar ataxia. PLoS One. 2013;8(12):e81884.
Shi CH, Schisler JC, Rubel CE, Tan S, Song B, McDonough H, et al. Ataxia and hypogonadism caused by the loss of ubiquitin ligase activity of the U box protein CHIP. Hum Mol Genet. 2014;23(4):1013–24.
Cordoba M, Rodriguez-Quiroga S, Gatto EM, Alurralde A, Kauffman MA. Ataxia plus myoclonus in a 23-year-old patient due to STUB1 mutations. Neurology. 2014;83(3):287–8.
Synofzik M, Schüle R, Schulze M, Gburek-Augustat J, Schweizer R, Schirmacher A, et al. Phenotype and frequency of STUB1 mutations: next-generation screenings in Caucasian ataxia and spastic paraplegia cohorts. Orphanet J Rare Dis. 2014;9:57.
Hayer SN, Deconinck T, Bender B, Smets K, Züchner S, Reich S, et al. STUB1/CHIP mutations cause Gordon Holmes syndrome as part of a widespread multisystemic neurodegeneration: evidence from four novel mutations. Orphanet J Rare Dis. 2017;12(1):31.
Gazulla J, Izquierdo-Alvarez S, Sierra-Martínez E, Marta-Moreno ME, Alvarez S. Inaugural cognitive decline, late disease onset and novel STUB1 variants in SCAR16. Neurol Sci. 2018;39(12):2231–3.
Genis D, Ortega-Cubero S, San Nicolás H, Corral J, Gardenyes J, de Jorge L, et al. Heterozygous STUB1 mutation causes familial ataxia with cognitive affective syndrome (SCA48). Neurology. 2018;91(21):1988–98.
De Michele G, Lieto M, Galatolo D, Salvatore E, Cocozza S, Barghigiani M, et al. Spinocerebellar ataxia 48 presenting with ataxia associated with cognitive, psychiatric, and extrapyramidal features: a report of two Italian families. Parkinsonism Relat Disord. 2019;65:91–6.
Ballinger CA, Connell P, Wu Y, Hu Z, Thompson LJ, Yin LY, et al. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol. 1999;19(6):4535–45.
Connell P, Ballinger CA, Jiang J, Wu Y, Thompson LJ, Höhfeld J, et al. The co-chaperone CHIP regulates protein triage decisions mediated by heat-shock proteins. Nat Cell Biol. 2001;3(1):93–6.
Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Höhfeld J, et al. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J Biol Chem. 2001;276(46):42938–44.
Qian SB, McDonough H, Boellmann F, Cyr DM, Patterson C. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–5.
Schmitz-Hubsch T, du Montcel ST, Baliko L, Berciano J, Boesch S, Depondt C, et al. Scale for the assessment and rating of ataxia: development of a new clinical scale. Neurology. 2006;66:1717–20.
Lee YC, Liao YC, Wang PS, Lee IH, Lin KP, Soong BW. Comparison of cerebellar ataxias: a three-year prospective longitudinal assessment. Mov Disord. 2011;26:2081–7.
Jacobi H, Rakowicz M, Rola R, Fancellu R, Mariotti C, Charles P, et al. Inventory of Non-Ataxia Signs (INAS): validation of a new clinical assessment instrument. Cerebellum. 2013;12(3):418–28.
Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”. A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–98.
Lin KN, Wang PN, Liu CY, Chen WT, Lee YC, Liu HC. Cutoff scores of the cognitive abilities screening instrument, Chinese version in screening of dementia. Dement Geriatr Cogn Disord. 2002;14(4):176–82.
Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2.
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9.
Kircher M, Witten DM, Jain P, O'Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5.
The Uniprot Consortium. Activities at the universal protein resource (UniProt). Nucleic Acids Res. 2014;42:D191–8.
Depondt C, Donatello S, Simonis N, Rai M, van Heurck R, Abramowicz M, et al. Autosomal recessive cerebellar ataxia of adult onset due to STUB1 mutations. Neurology. 2014;82(19):1749–50.
Sun M, Johnson AK, Nelakuditi V, Guidugli L, Fischer D, Arndt K, et al. Targeted exome analysis identifies the genetic basis of disease in over 50% of patients with a wide range of ataxia-related phenotypes. Genet Med. 2019;21(1):195–206.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–24.
Harding AE. Clinical features and classification of inherited ataxias. Adv Neurol. 1993;61:1–14.
Madrigal SC, McNeil Z, Sanchez-Hodge R, Shi CH, Patterson C, Scaglione KM, et al. Changes in protein function underlie the disease spectrum in patients with CHIP mutations. J Biol Chem. 2019;294:19236–45.
Funding
This work was supported by research grants from the Ministry of Science and Technology (MOST), Taiwan, to B.W.S. (103-2314-B-010-049-MY3, 104-2745-B-010-004, 106-2321-B-010-010, 107-2314-B-010-017). This work was also financially supported by the Brain Research Center, National Yang-Ming University from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan.
Author information
Authors and Affiliations
Contributions
Y.C. Lee and B.W.S. conceived and designed the study. H.H.C., C.T.H., and B.W.S. collected patient information and executed clinical analyses. Y.S.T. performed the mutational and bioinformatics analysis. H.H.C. and Y.C. Lee drafted the manuscript. C.T.H, Y.C. Liao, Y.C. Lee, and B.W.S. revised the manuscript. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chiu, HH., Hsaio, CT., Tsai, YS. et al. Clinical and Genetic Characterization of Autosomal Recessive Spinocerebellar Ataxia Type 16 (SCAR16) in Taiwan. Cerebellum 19, 544–549 (2020). https://doi.org/10.1007/s12311-020-01136-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-020-01136-4