Abstract
Immunoglobulin Kappa light chain gene (IGK) rearrangements are important complementary PCR targets for clonality assessment in mature B-cell proliferations in which detection of clonal VH-JH gene rearrangements fails. Failure to amplify VH-JH rearrangements is mainly attributable to somatic hypermutation. Detection of clonal IGK rearrangements is most relevant in the diagnosis of germinal center (GC) and post-GC B-cell lymphomas that are under a heavy somatic mutation process. Here we present three cases in which histology and immunophenotype of the tissue samples at time of diagnosis were inconclusive. PCR-based B-cell clonality assessment was performed using the standardized BIOMED-2 multiplex PCR protocols and Genescan analysis. No clonal VH-JH rearrangements were observed. Detection of a clonal IGK product in each of the three suspected cases supported the clonal origin of the B-cell proliferations. Interestingly, only rearrangements involving the Kde element were detected. Based in the clonal IGK-PCR result, a definitive diagnosis of follicular lymphoma (case 1 and 3) and extranodal marginal zone lymphoma (case 2) was established.
Similar content being viewed by others
Introduction
Analysis of immunoglobulin (Ig) gene rearrangements is a powerful tool to assess clonality in suspected B-cell proliferations, for which histological and immunophenotypic analysis is inconclusive. Since all cells from a B-cell malignancy derive from a single neoplastic transformed cell, they harbor the same Ig gene rearrangement pattern. Detection of this clonal pattern by polymerase chain reaction (PCR) amplification is a routine molecular methodology used in diagnostic hemathology.
Immunoglobulin heavy chain gene (IGH) rearrangements have been the most frequently used PCR targets in clonality assessment of mature B-cell proliferations at time of diagnosis [1–4]. However, false negative results do occur at a frequency that is mostly dependent on the IGH-PCR strategy and the lymphoma type and is related to the degree of somatic hypermutation (SHM) [5, 6]. SHM occurs in germinal center (GC) cells on the secondary lymphoid organs when the Ig on the B-cell membrane engages antigen. During this process, single-nucleotide mutations and to a less extent, insertions, or deletions of nucleotides, can occur in the entire VDJ exon of the IGH genes. These mutations alter VH and JH germinal sequences and can prevent primer annealing. Since the majority of the B-cell lymphomas arise from GC and post-GC cells they are somatically mutated and this is probably the main reason for the false negative IGH-PCR results observed in these types of lymphomas.
Incorporating alternative targets, such as the immunoglobulin Kappa light chain gene (IGK) segments, has been shown to improve clonality detection rates in mature B-cell malignancies that are heavily somatically mutated [7–10]. In principle, all mature B-cell malignancies will have rearranged their IGK genes. Rearrangements of the IGK genes start early during B-cell differentiation, soon after IGH genes, and follow a hierarchical order. A functional IGK rearrangement will generate an IgK+ B cell. Alternatively, inactivation of the IGK allele can take place through recombination of the kappa-deleting element (Kde) followed by rearrangement of the immunoglobulin lambda light chain (IGL) gene and giving rise to an IgL+ B lymphocyte. Accordingly, all IgK+ B-cell lymphomas should possess a clonal VK-JK rearrangement and in the vast majority of the IgL+ B-cell malignancies at least one clonal Kde rearrangement should be detected [11, 12].
Early PCR studies detecting IGK rearrangements in mature B-cell proliferations have evaluated mainly functional VK-JK rearrangements and, to a less extent, rearrangements involving de Kde segment [10, 13]. Multiplex primers and PCR protocols to detect IGK rearrangements were standardized during the BIOMED-2 Concerted Action CT98-3936 [14] and a guideline for efficient clonality testing in suspected B-cell neoplasms, with an inconclusive diagnosis or unusual histology or immunophenotype, was proposed [15]. Here we demonstrate how PCR analysis of IGK rearrangements contributes to confirm the presence of clonality in three illustrative cases where histology and immunophenotype did not allow the establishment of a definitive diagnosis and IGH-PCR amplification failed.
Pathology review and molecular analysis of three illustrative cases
Case 1
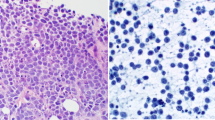
Case 1 concerns a lymph node (LN) from a patient with suspected non-Hodgkin lymphoma (NHL). After hematoxilin–eosin staining the histology of the LN showed a follicular pattern (Fig. 1). The follicles were small and atrophic, populated mainly by large cells. There was enlargement of the interfollicular region mainly due to the presence of a polymorphic population composed by few large B-cells, plasma cells, and small T cells. There were very few CD10+ cells in the germinal centers and none outside. BCL-6 was negative, BCL-2 staining was negative in the germinal centers, and Kappa and Lambda staining were inconclusive. There were no detectable BCL-2 rearrangements after FISH analysis (BCL-2 FISH DNA Probe, Split Signal, DAKO Denmark A/S, DK) in the tissue slides. Since a morphological diagnosis was not clear and there were no phenotypic aberrations, PCR-based clonality analysis of Ig gene rearrangements was conducted. The estimated percentage of tumor cell content in morphological analysis was greater than 80%.
Case 1: lymph node biopsy from a patient with suspected B-cell lymphoma. The hematoxilin–eosin (HE) staining shows a follicular pattern with small and atrophic follicles. The interfollicular region is enlarged due to the presence of a polymorphic population composed of few large B cells, plasma cells, and small T cells. There are very few CD10+ cells in the germinal centers and none outside. There was no evidence of Human Herpes Virus 8 (HHV8) infection. Bcl-2 and Bcl-6 staining were negative and Kappa and Lambda staining were inconclusive. There were no detectable BCL-2 rearrangements after FISH analysis (BCL-2 FISH DNA Probe, Split Signal, DAKO) in the tissue slides
For molecular analysis, DNA was extracted from the formalin fixed paraffin-embedded (FFPE) tissue using a proteinase K treatment and ethanol precipitation standard method. Quality of the sample DNA was assessed using the BIOMED-2 control gene primer set [14] and showed amplification of at least 300 base pairs (bp) amplicons. Two DNA quantities were used for PCR clonality analysis, namely 100 and 250 ng. The clonality profile of this patient is presented in Fig. 2. IGH VH-JH amplification showed neither clonal nor polyclonal signals in the FR1, FR2, and FR3 PCR tubes.
Case 1: immunoglobulin clonality profiles obtained in a lymph node with suspected B-cell lymphoma. Genescan profiles of IGH gene rearrangements using BIOMED-2 IGH-FR1, -FR2, and -FR3 tubes (a,b) and IGK-VK-JK (c) and IGK-Kde rearrangement tubes (d). Genescan profiles were obtained in an ABI 3130 platform (Applied Biosystems) using Genemapper (version 4.0) software (ABI prism). PCR reactions were performed in duplicate (100 and 250 ng DNA/PCR), with a monoclonal control (cell line) and a polyclonal control sample (tonsil). No detectable VH-JH rearrangements are seen in FR1, FR2, and FR3 PCR reactions, using neither 100 nor 250 ng of DNA (a,b). In tube IGK-A, polyclonal VK-JK rearrangements are seen (c). The clonal nature of the B-cell proliferation in case 1 is supported by the presence of two clonal Kde rearrangements (arrows), in IGK-tube B: 283 bp and 381 bp (d). Note that the 381 bp clonal peak (d) is less intense, probably reflecting the degradation of the extracted DNA. Also, IGH-DH1-6-JH (tube D) rearrangements were polyclonal (data not shown).
IGH-PCR failure should be interpreted carefully, mainly when FFPE DNA is used, as in the present analysis. Absence of both clonal and polyclonal VH-JH rearrangements was not expected since the estimated percentage of suspect B cells was 80%, far above the analytical sensitivity of this PCR approach [14]. Also, a similar negative result was obtained using either 100 or 250 ng of DNA, precluding the presence of PCR inhibitors resulting from the DNA extraction procedure. Insufficient DNA quality could be the potential source of this false negative result. However, PCR assessment of the DNA quality resulted in amplicons of up to 300 bp. This implies mild fragmentation of the DNA and potential lack of amplification of clonal rearrangements larger than 300 bp. The expected size range of the BIOMED-2 VH-JH PCR products varies between 100–170, 250–295, and 310–360 bp for FR3, FR2, and FR1, respectively. Thus, at least FR2 and FR3 fragments should have been amplified.
A polyclonal pattern was seen in tube IGK-A but two clonal peaks, of 283 and 381 bp each, were obtained in tube IGK-B, supporting the clonal nature of the B-cell lymphoproliferation (Fig. 2). The second peak of 381 bp obtained in both IGK-B replicates is faint in contrast to the 283 bp, probably due to degradation of the purified DNA. Thus, the presence of a clonal IGK pattern, together with the suspicious morphology and in the absence of phenotypic aberrations, contributed to establish the diagnosis of follicular lymphoma (FL).
Case 2
Case 2 concerns a patient with a skin lesion in the nasal region. Histological examination suggested a low grade B-cell lymphoma (Fig. 3). A polymorphous CD20+, CD10−, CD23−, CD5−, and CD3− population was observed. Kappa and Lambda staining failed to show a monotypic population. To discriminate between a reactive condition and a clonal B-cell proliferation, molecular analysis of Ig gene rearrangements was performed. The estimated percentage of B cells was 50% of which most were suspected of being malignant.
Case 2: skin lesion biopsy from the nasal region. Histology suggests a low grade B-cell lymphoma. A polymorphous CD20+, CD10−, CD23−, CD5−, and CD3− population was observed. However, Kappa and Lambda staining failed to show a monotypic population. The estimated percentage of B cells is 50% of which most were suspected of being malignant B cells
DNA was extracted from the snap-frozen tissue of the same lesion using Gentra Puregene Cell and Tissue kit (QIAGEN, Maryland, USA). Isolated DNA showed amplification up to 600 bp of the BIOMED-2 control gene PCR. Each PCR reaction was done in duplicate, with 100 ng of input DNA. The Ig Genescan profiles of this patient are presented in Fig. 4. IGH VH-JH amplification gave clear polyclonal signals in FR1, FR2, and FR3 tubes, suggesting the presence of a polyclonal B-cell proliferation. However, a clear clonal 238 bp peak was obtained in IGK-B tube, supporting the clonal nature of the skin lesion. Interestingly, there were no detectable clonal VK-JK gene rearrangements. In this case, with an unusual histological pattern and no phenotypical alterations, the presence of a clonal Kde rearrangement contributed to the diagnosis of extranodal marginal zone lymphoma (MZL).
Case 2: immunoglobulin gene rearrangement results obtained in a skin lesion with a suspected B-cell lymphoproliferation. Genescan profiles are shown after amplification of IGH FR1, FR2, and FR3 (a,b) and IGK-VK-JK (c) and IGK-Kde rearrangements (d) , using BIOMED-2 primers and protocols. Genescan was performed in a ABI 3130 platform, using Genemapper (version 4.0) software (ABI prism). Genescan results of the patient’s DNA are shown in duplicate (100 ng input DNA in each PCR), and a monoclonal (cell line) and polyclonal control (tonsil) DNA sample. No clonal VH-JH gene rearrangements were detected in any of the 3 PCR tubes: FR1, FR2, and FR3 IGH-PCR tubes gave clear polyclonal signals (a,b). IGK-A tube shows polyclonal VK-JK rearrangements (c). A clear monoclonal peak with 238 bp (arrow) is seen in IGK-B tube, supporting the clonal origin of the skin lesion (d). Also, incomplete IGH rearrangements in IGH-D and -E tubes were in polyclonal configuration (data not shown)
Case 3
In case 3, a cervical lymph node was obtained from a patient with suspected lymphoma. The histology of the biopsy showed intrasinusoidal infiltration by predominantly small lymphoma cells (Fig. 5). The suspected cells were CD20+, Pax-5+, CD10+, BCL-6+, BCL-2+, CD5−, and CD30− by immunostaining, suggesting follicular lymphoma with intrasinusoidal infiltration. However, the percentage of suspected cells was no greater than 15%. In order to demonstrate their clonal origin, Ig gene rearrangement analysis was performed.
Case 3: cervical lymph node biopsy from a patient with suspected lymphoma. The histology of the biopsy combined with the immunostainings (CD20+, Pax-5+, CD10+, BCL-6+, BCL-2+, CD5−, CD30−) suggested a follicular lymphoma with intrasinusoidal infiltration. The percentage of suspected cells was no greater than 15% and there was no phenotypical evidence of clonality
DNA was extracted from the snap frozen biopsy using EZ1 DNA Tissue Kit (QIAGEN). The control gene PCR showed faint amplification of the 300 bp amplicon suggesting poor preservation of the purified DNA. Clonality of the suspected B-cell proliferation was supported by the presence of two clonal peaks in tube IGK-B, with 237 and 307 bp, each (Fig. 6). Polyclonal patterns were obtained in IGH tubes A-C, being more prominent in FR1 and FR3 than FR2 amplifications and suggesting the presence of reactive B cells. Again, no clonal VK-JK rearrangements could be detected (Fig. 6). In this case, with an aberrant histology but with no phenotypical evidence of clonality, the diagnosis of FL was supported by the presence of a clonal Kde pattern.
Case 3: immunoglobulin clonality profiles obtained in the lymph node of a patient with suspected lymphoma. Genescan profiles of IGH gene rearrangements using BIOMED-2 IGH FR1, FR2, and FR3 PCR tubes (a) and of IGK-VK-JK and IGK-Kde rearrangement tubes (b). Genescan profiles were obtained in an ABI 3130 platform (Applied Biosystems) using Genemapper (version 4.1) software (ABI prism). DNA was extracted from the snap frozen lymph node biopsy (Case 3). Genescan profiles are shown in duplicate (100 ng of sample’s DNA in each PCR) and a monoclonal (cell line) and polyclonal (tonsil) DNA control were included. A polyclonal pattern is seen in IGH FR1, FR2, and FR3 tubes (a). VK-JK rearrangements were in polyclonal configuration (b). The clonal origin of the B-cell proliferation is supported by the two clonal Kde rearrangements (arrows) seen in IGK tube B (c).
Discussion
-
1.
False negative IGH-PCR results
PCR-based IGH analysis failed to demonstrate clonality in the three cases presented above. In the first case, no specific IGH-PCR amplification products were obtained at all. In case 2 and 3, only polyclonal VH-JH rearrangements were seen in the three IGH-PCR tubes due to the presence of reactive polyclonal B cells.
Failure in detecting clonal VH-JH rearrangements in mature B-cell lymphomas is mainly attributable to the presence of somatic hypermutation [14]. In the two types of lymphoma presented above, FL and extranodal MZL, the IGH genes are expected to be highly mutaded, according to their origin in GC and/or post-GC cells. Moreover, in FL ongoing somatic mutations are known to occur. Thus, failure to detect VH-JH rearrangements in the three analyzed cases was not totally unexpected.
Higher false negative VH-JH PCR results have been reported in disease categories traditionally associated with high levels of SHM such as diffuse large B-cell lymphomas (DLBCL), MZL, and FL [9, 16, 17]. During the germinal center reaction, SHM targets the entire rearranged VDJ exon of the IGH gene but its activity is greatest in and around the CDR3 [18]. Accordingly, all FRs are affected but the FR3 is the most vulnerable region involved in the mutation process. In these types of lymphomas, PCR strategies focusing on FR1 and FR2 sequences result in higher clonality detection rates than on FR3 sequences [1, 6, 9, 16]. Additionally, SHM or excessive exonucleolytic activity can also target the FR4 region, located on the JH segment. This would impair annealing of the JH consensus primer and there would be no amplification with FR1, FR2 and FR3 IGH-PCR tubes [14]. Most probably, this was the cause for the negative results obtained in case 2 and 3.
On going from FR3 to FR2 and FR1, the size of the obtained PCR fragments increase. This is not a limitation when testing DNA obtained from fresh/frozen tissues. However, when analyzing poor quality DNA, the FR1-PCR may not be informative, in particular if the control gene PCR is smaller than ∼300 bp [14, 19]. This was probably the reason why no IGH amplification was obtained in case 1, a highly mutated FL. When analyzing DNA obtained from FFPE mutated lymphomas, the smaller FR2 and FR3 amplicons, being more prone to mutations, are usually less informative. This discrepancy is more evident in GC and post-GC lymphomas where overall VH-JH detection rates are inferior to those obtained in pre-GC lymphomas [14, 17, 20, 21]. Importantly, amplification of FR3, FR2, and FR1 can fail [19, 21] despite amplification of ∼300 bp control gene PCR.
IGH false negative results in mature B-cell malignancies can be explained by other reasons. The use of FR1, FR2, and FR3 multiplex PCR primers does not detect incomplete DH-JH rearrangements. Incomplete IGH rearrangements are rarely seen as an isolated clonal marker in highly mutated lymphomas [9]. However, they can still be informative in the presence of SHM [22], when both VH-JH and IGK-PCR fail [15].
Another explanation for IGH-PCR failure is the presence of chromosomal translocations that impair one allele from annealing to the PCR primers [23]. Chromosomal translocations are present in many GC and post-GC lymphomas such as the t(14;18), which is characteristic of FL. This explanation cannot be accounted for in case 1 since no BCL-2 rearrangements were detected by FISH. In case 3, detection of the t(14;18) was not carried on, so the presence of this rearrangement, rendering one allele unsuitable for amplification, cannot be excluded.
Finally, unexpected IGH rearrangements of smaller or larger size can also occur in mutated lymphomas due to large deletions in VH segments or annealing of the JHc primer to a downstream JH segment, respectively [24, 25]. If misinterpreted, these rearrangements can escape detection in routine clonality analysis and contribute to false negative clonality results.
-
2.
Added value of IGK-PCR analysis
In the three suspected lymphoproliferations studied, IGK-PCR was able to detect clonality whereas IGH-PCR analysis failed. In total, five clonal IGK rearrangements were amplified: two Kde rearrangements in case 1 and 3, and one Kde rearrangement in case 2.
The multiplex IGK-PCR strategy used [14] detects both VK-JK and rearrangements involving the Kde element. This Kde element can rearrange with either a VK segment or an intronic recombination signal sequence (intron-RSS) lying between the JK and CK segments. Thus, in the same allele a VK-JK and a Kde rearrangement can be detected. This results in four amplifiable IGK rearrangements per cell [14] contrasting to the only two amplifiable VH-JH rearrangements. In the three analyzed B-cell proliferations, the potential amplification of rearrangements involving the Kde element largely contributed to demonstrate their clonal origin. Interestingly, in case 1, as well as in case 3, the presence of a biallelic clonal Kde rearrangement strongly suggests the expression of an IgL chain. Importantly, all clonal PCR products were obtained in duplicate PCR reactions, confirming the presence of true clonal rearrangements [9]. In agreement to these findings, the estimated percentage of malignant lymphocytes in each case (80%, 50%, and 15% in case 1, 2, and 3, respectively) was above the detection limit of the used IGK-PCR strategy [14] precluding amplification of restricted Ig gene rearrangements and pseudoclonality. Pseudoclonality or false positivity is more frequent in tissue samples in which there are few lymphocytes [23]. Thus, the estimated number of suspected and normal (reactive) lymphocytes in the tissue sample should always be taken into account for a correct interpretation of the PCR results.
Detection of IGK rearrangements has been highlighted in clonality assessment of GC and post-GC lymphomas [9, 16, 21]. In these highly mutated lymphomas, IGK-PCR can support the clonal origin of the B-cell proliferation when IGH-PCR fails [8, 10, 26–28]. In fact, functional VK-JK rearrangements are less prone to somatic hypermutation than IGH rearrangements [29, 30] and thus, more suitable to be amplified in somatically mutated lymphomas. Moreover, inactivating Kde rearrangements, due to deletion of the intronic JK-CK region, remove the IGK enhancer, which is essential for the somatic hypermutation process to occur. Accordingly, Kde rearrangements are always unmutated [14] and easily amplified. Interestingly, in the three cases presented above, rearrangements involving the Kde segment were the only type of clonal IGK rearrangements detected. This is in agreement with the expected heavily mutated status of the IG genes in FL and extra MZL. Thus, rearrangements involving a non-mutated Kde segment were preferentially amplified.
The added value of IGK-PCR in FFPE GC and post-GC lymphomas has been reported by a number of studies [7, 13, 21, 31]. Some studies do report an even higher sensitivity for IGK assays comparing to IGH-PCR, in FFPE tissues [17, 32]. The majority of IGK-PCR amplicons is smaller than IGH-FR1 PCR products and thus is more appropriate for poor quality DNA. In particularly, the small product size of the most frequently amplified VK gene family, VKf1, (∼150 bp in tube A and ∼250 bp in tube B) facilitates amplification. In case 1, due to the poor quality of DNA, successful amplification of IGK rearrangements, in contrast to VH-JH rearrangements, was not unexpected. Interestingly, also in case 1, a faint PCR amplicon of 381 bp (Fig. 2) was detected although the control gene PCR was smaller than 300 bp. This fragment of 381 bp corresponds to a Kde rearrangement with either a VKfII/VK4 or VK5 segment [14] and in combination with the 283 amplicon contributes to confirm clonality.
-
3.
General conclusion
In the three clinically suspected cases described above, histology and immunophenotype of the biopsy tissues were not conclusive. In order to establish a final diagnosis clonality was assessed using IGH- and IGK-PCR. PCR detection of clonal Kde rearrangements in all cases supported the diagnosis of follicular lymphoma (case 1 and 3) and extranodal marginal zone lymphoma (case 2). In case 1, a subsequent biopsy revealed a FL grade 3b. The three cases presented illustrate the utility of analyzing IGK gene rearrangements in pathology specimens in which there is a strong clinical and histological suspicion of malignant lymphoma but with no detectable clonal IGH rearrangements. PCR amplification of IGK rearrangements has been found particularly useful in clonality assessment of lymphoproliferative disorders that arise from GC and post-GC cells such as MZL, FL, and DLBCL, in contrast to pre-GC lymphomas. However, not all LN or other biopsy tissues have diagnostic problems, with the percentage of cases in which PCR is performed being dependent on the experience of the hematopathology laboratory. Importantly, clonality assessment based on PCR also depends on the availability of appropriate archival material. In FFPE DNA samples, IGK PCR is probably more informative than IGH amplification mainly when highly mutated lymphomas such as FL are analyzed.
Obtaining a clonal IGK-PCR result in the absence of an IGH amplification supports the clonal origin of suspected B-cell proliferations. However, molecular data interpreted in isolation can be misleading. False-negative PCR results can occur. Moreover, presence of clonal PCR results does not always imply malignancy, and false positivity can happen. To avoid misinterpretation of molecular data, technical and immunological pitfalls should be carefully evaluated between the pathologist and the molecular biologist. Establishment of a definite diagnosis should only be accomplished after integration of molecular results with histological and immunophenotypical data.
References
Aubin J, Davi F, Nguyen-Salomon F, Leboeuf D, Debert C, Taher M, Valensi F, Canioni D, Brousse N, Varet B et al (1995) Description of a novel FR1 IgH PCR strategy and its comparison with three other strategies for the detection of clonality in B cell malignancies. Leukemia 9:471–479
Diss TC, Peng H, Wotherspoon AC, Isaacson PG, Pan L (1993) Detection of monoclonality in low-grade B-cell lymphomas using the polymerase chain reaction is dependent on primer selection and lymphoma type. J Pathol 169:291–295
McCarthy KP, Sloane JP, Wiedemann LM (1990) Rapid method for distinguishing clonal from polyclonal B cell populations in surgical biopsy specimens. J Clin Pathol 43:429–432
Trainor KJ, Brisco MJ, Wan JH, Neoh S, Grist S, Morley AA (1991) Gene rearrangement in B- and T-lymphoproliferative disease detected by the polymerase chain reaction. Blood 78:192–196
Ben-Ezra J (1992) Variable rate of detection of immunoglobulin heavy chain V-D-J rearrangement by PCR: a systematic study of 41 B-cell non-Hodgkin’s lymphomas and leukemias. Leuk Lymphoma 7:289–295
Derksen PW, Langerak AW, Kerkhof E, Wolvers-Tettero IL, Boor PP, Mulder AH, Vrints LW, Coebergh JW, van Krieken JH, Schuuring E, Kluin PM, van Dongen JJ (1999) Comparison of different polymerase chain reaction-based approaches for clonality assessment of immunoglobulin heavy-chain gene rearrangements in B-cell neoplasia. Mod Pathol 12:794–805
Amara K, Trimeche M, Ziadi S, Sriha B, Mokni M, Korbi S (2006) PCR-based clonality analysis of B-cell lymphomas in paraffin-embedded tissues: Diagnostic value of immunoglobulin [kappa] and [lambda] light chain gene rearrangement investigation. Pathol Res Pract 202:425–431
Diss TC, Liu HX, Du MQ, Isaacson PG (2002) Improvements to B cell clonality analysis using PCR amplification of immunoglobulin light chain genes. Mol Pathol 55:98–101
Evans PAS, Pott C, Groenen PJTA, Salles G, Davi F, Berger F, Garcia JF, van Krieken JHJ M, Pals S, Kluin P, Schuuring E, Spaargaren M, Boone E, Gonzalez D, Martinez B, Villuendas R, Gameiro P, Diss TC, Mills K, Morgan GJ, Carter GI, Milner BJ, Pearson D, Hummel M, Jung W, Ott M, Canioni D, Beldjord K, Bastard C. Delfau-Larue MH, van Dongen JJM, Molina TJ, and Cabecadas J (2006) Significantly improved PCR-based clonality testing in B-cell malignancies by use of multiple immunoglobulin gene targets. Report of the BIOMED-2 Concerted Action BHM4-CT98-3936 Leukemia 21: 207–214.
Kuppers R, Zhao M, Rajewsky K, Hansmann ML (1993) Detection of clonal B cell populations in paraffin-embedded tissues by polymerase chain reaction. Am J Pathol 143:230–239
Langerak AW, Nadel B, De Torbal A, Wolvers-Tettero IL, van Gastel-Mol EJ, Verhaaf B, Jager U, van Dongen JJ (2004) Unraveling the consecutive recombination events in the human IGK locus. J Immunol 173:3878–3888
van der Burg M, Tumkaya T, Boerma M, de Bruin-Versteeg S, Langerak AW, van Dongen JJ (2001) Ordered recombination of immunoglobulin light chain genes occurs at the IGK locus but seems less strict at the IGL locus. Blood 97:1001–1008
Pai RK, Chakerian AE, Binder JM, Amin M, Viswanatha DS (2005) B-cell clonality determination using an immunoglobulin [kappa] light chain polymerase chain reaction method. J Mol Diagn 7:300–307
van Dongen JJM, Langerak AW, BruggemannM EPAS, Hummel M, Lavender FL, Delabesse E, Davi F, Schuuring E, Garcia-Sanz R, van Krieken JHJM, Droese J, Gonzalez D, Bastard C, White HE, Spaargaren M, Gonzalez M, Parreira A, SmithJL MGJ, Kneba M, Macintyre EA (2003) Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene recombinations in suspect lymphoproliferations: Report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia 17:2257–2317
van Krieken JHJM, Langerak AW, Macintyre EA, Kneba M, Hodges E, Sanz RG, Morgan GJ, Parreira A, Molina TJ, Cabecadas J, Gaulard P, Jasani B, Garcia JF, Ott M, Hannsmann ML, Berger F, Hummel M, Davi F, Bruggemann M, Lavender FL, Schuuring E, Evans PAS, White H, Salles G, Groenen PJTA, Gameiro P, Pott C, Dongen JMV (2006) Improved reliability of lymphoma diagnostics via PCR-based clonality testing: Report of the BIOMED-2 Concerted Action BHM4-CT98-3936. Leukemia 21:201–206
Catherwood MA, Gonzalez D, Patton C, Dobbin E, Venkatraman L, Alexander HD (2007) Improved clonality assessment in germinal centre/post-germinal centre non-Hodgkin lymphomas with high rates of somatic hypermutation. J Clin Pathol 60:524–528
McClure RF, Kaur P, Pagel E, Ouillette PD, Holtegaard CE, Treptow CL, Kurtin PJ (2006) Validation of immunoglobulin gene rearrangement detection by PCR using commercially available BIOMED-2 primers. Leukemia 20:176–179
Papavasiliou FN, Schatz DG (2002) Somatic hypermutation of immunoglobulin genes: merging mechanisms for genetic diversity. Cell 109:S35
Hebeda K, Van Altena M, Rombout P, Van Krieken J, Groenen P (2009) PCR clonality detection in Hodgkin lymphoma. J Hematopathol 2:34–41
Bagg A, Braziel RM, Arber DA, Bijwaard KE, Chu AY (2002) Immunoglobulin heavy chain gene analysis in lymphomas: a multi-center study demonstrating the heterogeneity of performance of polymerase chain reaction assays. J Mol Diagn 4:81–89
Liu H, Bench AJ, Bacon CM, Payne K, Huang Y, Scott MA, Erber WN, Grant JW, Du MQ (2007) A practical strategy for the routine use of BIOMED-2 PCR assays for detection of B- and T-cell clonality in diagnostic haematopathology. Br J Haematol 138:31–43
Gonzalez D, Balanzategui A, Garcia-Sanz R, Gutierrez N, Seabra C, van Dongen JJ, Gonzalez M, San Miguel JF (2003) Incomplete DJH rearrangements of the IgH gene are frequent in multiple myeloma patients: immunobiological characteristics and clinical implications. Leukemia 17:1398–1403
Hoeve MA, Krol AD, Philippo K, Derksen PW, Veenendaal RA, Schuuring E, Kluin PM, van Krieken JH (2000) Limitations of clonality analysis of B cell proliferations using CDR3 polymerase chain reaction. Mol Pathol 53:194–200
Langerak AW (2008) Undersized, oversized? It is not one-size-fits-all in lymphoid clonality detection. Leuk Res 32:203–204
Vargas RL, Felgar RE, Rothberg PG (2008) Detection of clonality in lymphoproliferations using PCR of the antigen receptor genes: does size matter? Leuk Res 32:335–338
Catherwood MA, Alexander HD, McManus DT, Cuthbert RJ, Morris TC (2003) Immunoglobulin gene rearrangement investigations in the diagnosis of lymphoid malignancies from formaldehyde-fixed biopsies. Leuk Lymphoma 44:645–648
Gong JZ, Zheng S, Chiarle R, De Wolf-Peeters C, Palestro G, Frizzera G, Inghimari G (1999) Detection of immunoglobulin kappa light chain rearrangements by polymerase chain reaction. An improved method for detecting clonal B-cell lymphoproliferative disorders. Am J Pathol 155:355–363
Sandberg Y, van Gastel-Mol EJ, Verhaaf B, Lam KH, van Dongen JJ, Langerak AW (2005) BIOMED-2 multiplex immunoglobulin/T-cell receptor polymerase chain reaction protocols can reliably replace Southern blot analysis in routine clonality diagnostics. J Mol Diagn 7:495–503
Kosmas C, Stamatopoulos K, Papadaki T, Belessi C, Yataganas X, Anagnostou D, Loukopoulos D (1998) Somatic hypermutation of immunoglobulin variable region genes: focus on follicular lymphoma and multiple myeloma. Immunol Rev 162:281–292
Stamatopoulos K, Kosmas C, Papadaki T, Pouliou E, Belessi C, Afendaki S, Anagnostou D, Loukopoulos D (1997) Follicular lymphoma immunoglobulin kappa light chains are affected by the antigen selection process, but to a lesser degree than their partner heavy chains. Br J Haematol 96:132–146
Berget E, Helgeland L, Molven A, Vintermyr OK (2011) Detection of clonality in follicular lymphoma using formalin-fixed, paraffin-embedded tissue samples and BIOMED-2 immunoglobulin primers. J Clin Pathol 64:37–41
Halldorsdottir AM, Zehnbauer BA, Burack WR (2007) Application of BIOMED-2 clonality assays to formalin-fixed paraffin embedded follicular lymphoma specimens: superior performance of the IGK assays compared to IGH for suboptimal specimens. Leuk Lymphoma 48:1338–1343
Conflict of interest
The authors declare that they have no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Gameiro, P., Sebastião, M., Spetalen, S. et al. The added value of immunoglobulin Kappa light chain gene (IGK) rearrangement analysis in suspected B-cell lymphomas: three illustrative cases. J Hematopathol 5, 45–56 (2012). https://doi.org/10.1007/s12308-011-0132-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12308-011-0132-6