Abstract
Fusarium wilt caused by Fusarium oxysporum f. sp. lycopersici (Fol) is a major fungal disease of tomato (Solanum lycopersicum L.). Xylem sap protein 10 (XSP10) and Salicylic acid methyl transferase (SlSAMT) have been identified as putative negative regulatory genes associated with Fusarium wilt of tomato. Despite their importance as potential genes for developing Fusarium wilt disease tolerance, very little knowledge is available about their expression, cell biology, and functional genomics. Semi-quantitative and quantitative real-time PCR expression analysis of XSP10 and SlSAMT, in this study, revealed higher expression in root and flower tissue respectively in different tomato cultivars viz. Micro-Tom (MT), Arka Vikas (AV), and Arka Abhed (AA). Therefore, the highly up-regulated expression of XSP10 and SlSAMT in biotic stress susceptible tomato cultivar (AV) than a multiple disease resistant cultivar (AA) suggested the disease susceptibility nature of these genes for Fusarium wilt. Sub-cellular localization analysis through the expression of gateway cloning constructs in tomato protoplasts and seedlings showed the predominant localization of XSP10 in the nucleus and SlSAMT at the cytoplasm. A strong in vivo protein–protein interaction of XSP10 with SlSAMT at cytoplasm from bi-molecular fluorescent complementation study suggested that these two proteins function together in regulating responses to Fusarium wilt tolerance in tomato.
Similar content being viewed by others
Introduction
Tomato (Solanum lycopersicum. L) is grown worldwide as a major crop and consumed because of its high nutrition value and fibre content (Ranjan et al. 2012). Owing to vulnerability to abiotic and biotic stresses plants have developed various mechanisms to manage these stresses (Dresselhaus and Hückelhoven 2018). Fusarium wilt caused by fungal pathogen Fusarium oxysporum f. sp. lycopersici (Fol) has brought drastic negative impact by jeopardizing crop yield of tomato (Goswami and Kistler 2004). Fol enters the plant through the mechanistic route during injury, colonizes the apoplastic spaces of the root cortex, covers the stele and the xylem vascular tissues. This leads to clogging of vessels, yellowing of leaves, wilting, and finally cell death (Joshi, 2018). Upon infection, Fol secretes effector molecules to suppress host genes and defense-related proteins (Gawehns et al. 2015).
Xylem sap proteins (XSPs) belong to a unique non-specific lipid-binding protein (nsLTPs) family (Rep et al. 2003a). nsLTPs family contains a conserved motif with 8-cysteine residues which form intramolecular disulphide bonds (Rep et al. 2003a). The nsLTPs family protein, XSP10 represents a compatible protein required for Fol to develop complete Fusarium wilt disease (Krasikov et al. 2011). Further, the XSP10 is involved in trafficking essential lipids from intracellular membrane to pathogen resulting in disease susceptibility in tomato (Blein et al. 2002). Xylem sap protein is functionally characterized in Brassica oleracea (Ligat et al. 2011), Brassica napus (Luo and Zhang 2019), Cucumis sativus (Buhtz et al. 2004), Solanum lycopersicum (HOUTERMAN et al. 2007), Cucurbita maxima (Satoh 2006), Glycine max (Subramanian et al. 2009), Zea mays (Alvarez et al. 2008), Pyrus communis (Biles and Abeles 1991) and Gossypium hirsutum (Yang et al. 2019). Apart from XSP10 protein, one-dimensional electrophoresis data have shown a high accumulation of pathogenesis related proteins (PRPs), chitinase, peroxidase, and β-1,3, glucanases in Fol infected xylem tissues (Rep et al. 2003b).
Salicylate (SA) is a key precursor to methyl salicylate (MeSA) which is an important plant volatile substance that functionally activates plant-herbivory defense response (Tieman et al. 2010). MeSA is involved in local priming and systemic acquired resistance (SAR) (Liu et al. 2011). S-Adenosyl-l-methionine: salicylic acid carboxyl methyl transferase (SAMT) was isolated from petals of the annual California plant Clarkia breweri (Ross et al. 1999), Stephanotis floribunda (Pott et al. 2003), Antirrhinum majus (Snapdragon) (Negre et al. 2002), Arabidopsis thaliana, Hoya carnosa, and Petunia hybrid (Effmert et al. 2005). The structure of SAMT indicates that the presence of Trp residue in position 226 forms part of the salicylic acid-binding site in Clarkia brewerii (Zubieta et al. 2003). SAMT encodes a protein that releases MeSA as by-product (Ament et al. 2010). However, the involvement of SA and JA induces Fol disease susceptibility (Di et al. 2017). Recent experimental data shows SlSAMT-knockdown reduced susceptibility to virulent root invading fungus Fol (Ament et al. 2010). Constitutive expression of MeSA in Arabidopsis thaliana leads to disease susceptibility to the bacterial pathogen Pseudomonas syringae and fungal pathogen Golovinomyces orontii (Koo et al. 2007). In the transgenic Arabidopsis plant, over-expression of AtBSMT1 accumulated higher MeSA but did not develop SAR in pathogen infected leaves (Liu et al. 2010). It has been shown that over-expression of SlSAMT1 provides long term protection against Xanthomonas campestris (Xcv) infection (Tieman et al. 2010). It has also been demonstrated that MeSA and SA inhibit citrus canker caused by Xanthomonas citri (Lima Silva, 2019).
Despite their known significant role as putative disease susceptible genes of Fol, little is known about the expression levels, their cell biology, and protein partners of XSP10 and SlSAMT in cultivated tomato. Here, we performed a systematic expression study of XSP10 and SlSAMT genes in different tissues of tomato cultivars such as viz. Micro-Tom (MT), Arka Vikas (AV), and Arka Abhed (AA) for their tissue-specific expression, and to identify the most susceptible tomato cultivar associated with Fusarium wilt. Further, we performed transient in vivo cell biological analysis to understand the sub-cellular localization and identify strong protein–protein interacting partners of XSP10 and SlSAMT in tomato protoplast and seedlings. The current study provides knowledge on cell and molecular biology of two key Fusarium wilt responsive genes XSP10 and SlSAMT in tomato that can be explored to unravel their functional significance in developing Fusarium wilt tolerance.
Materials and methods
Plant materials and growth condition
Seeds of different tomato cultivars MT, AV, and AA were procured from ICAR-IIHR, Bangalore, India. The seeds were surface sterilized with 70% ethanol for 5 min, followed by washing with 4% sodium hypochlorite (W/V) for 10 min and thrice with distilled water. The sterile seeds were germinated in dark for 3 days in ½ MS (Murashige and Skoog) media supplemented with 3% sucrose and 0.3% Gelrite (Sigma Aldrich) followed by growing in the plant growth chamber at 25–28 °C,70% relative humidity, 16/8 h photoperiod with a light intensity of 150 µE m−2 s−2. Early grown 10–12 days old seedlings were then transferred to soil-rite in the ratio of 4:1:1 (cocopeat:perlite:vermiculite) for flowering and fruit development.
In silico analysis of XSP10 and SlSAMT
The information of nucleotide sequences of XSP10 and SlSAMT was accessed from NCBI and Sol-Genomics Network (https://solgenomics.net/). NCBI-BLAST search for identifying the homologs of XSP10 and SlSAMT proteins in the Solanaceae family was performed. The best hits aligned amino acid sequences thus obtained were imported into DNAMAN software (https://www.lynnon.com/) and a molecular phylogenetic tree was constructed by the neighbor-joining statistical method with 1000 bootstrap replicates. SMART online tools (http://smart.embl-heidelberg.de/) were used to predict the gene structure, domain architecture and graphically represented by IBS.1 software (Liu et al. 2015).
In silico predictions of localization and interactions
The protein sequences of XSP10 and SlSAMT were retrieved from BLASTP and imported into online software predictor CELLO.2.GO (Yu et al. 2014). The localizations were predicted according to the best hit score having user-specified threshold E-value 0.001 (default) (Yu et al. 2014).
In silico protein interaction prediction of XSP10 and SlSAMT with stress-associated proteins was carried out using STRING.11.0 tool (Szklarczyk et al. 2015). The amino acid sequences were uploaded in multiple sequence FASTA format and protein–protein networks (PPNs) were generated based on text mining, co-expression, and experimental database (Szklarczyk et al. 2015). The interaction correlation value was considered as strong (0.5–1), moderate (0.3–0.5), and weak (0.1–0.3) (Zhang and Zhang 2019). The nodes describe the protein encoded by a single gene and edges for the association of specific binding of proteins.
RNA isolation and cDNA synthesis
Tissues of the root, stem, leaf, flower, fruit, and seed of three tomato cultivars namely MT, AV, and AA were collected. Total RNA was extracted using E.N.Z.A ® Plant RNA kit, USA. 100 mg of each tissue was ground with liquid nitrogen for tissue rupture. About 40µL of RNAse –free water was added to get the final eluted product. A Prime Script™ RT reagent Kit with gDNA Eraser (Clontech, Takara) was used in synthesizing cDNA transcripts from each of 1 µg aliquots of non-denaturing total RNA.
Semi-quantitative PCR and qRT-PCR
The tomato cultivars used for the study were Micro-Tom (MT), Arka Vikas (AV) and Arka Abhed (AA). MT has been recognized as one of the model cultivars for tomato research as it shares some important advantages with Arabidopsis such as small size, short life cycle and suited for indoor cultivation (Masahito Shikata and Hiroshi Ezura 2016). AV is developed by ICAR-IIHR Bangalore which is tolerant to moisture stress but susceptible to wilt disease. Plants of AV are semi-determinate with dark green foliage suitable for cultivation in kharif/rabi seasons and matures in about 140 days (Upreti and Thomas 2015). AA is developed by ICAR-IIHR Bangalore which is resistant to multiple diseases such as tomato leaf curl (Ty2 + Ty3), bacterial wilt, early blight and late blight (Ph2 + Ph3). Plants of AV are semi-determinate with dark green foliage suitable for kharif/rabi cultivation and matures in about 150 days (Sunitha, 2020). Six different tissues namely root, stem, leaf, flower fruit, and the seed of each cultivar were tested for the expression of XSP10 and SlSAMT. The elongation factor α 1(EFα1) was used as an internal control in both RT-PCR and qRT-PCR. Semi-quantitative PCR of XSP10 and SlSAMT genes was performed by using the DNA polymerase EmraldAmp ® GT master mix (Takara Bio INC). For RT-PCR the thermocycling program was used as follows: 94°C for 5 min, 55°C for 45 s, 72°C for 15 s followed by 35 cycles. Using SYBR green fluorophores (Applied Biosystems), the q-PCR program was set as follows 95°C for 7 min, 60°C 30 s, for 40 cycles. The melting curve was observed at 60°C for 30 s (Applied Biosystems, USA). All data analyses were performed following the method (Livak and Schmittgen 2001). For statistical analysis, the experiment was performed with three independent biological replicates and each reaction was set up with three technical replicates. The experiment was repeated thrice. For setting control measurements with high CT (cycle threshold) levels were arbitrarily set to one. The parametric two sample t-test measurement was employed. Primers used in the study are given in Table S1.
Generation of Gateway sub-cellular localization constructs
The primers were designed for cloning open reading frames (ORFs) of XSP10 and SlSAMT gene flanking gateway adaptor sites attB1 in forward and attB2 sites in reverse orientation. The PCR product was cloned into the entry vector pDNR221 using BP Clonase II enzymes (Gateway ™ Technology, Invitrogen) (Gehl et al. 2009). Entry clones of XSP10 genes with oriented (+) stop codon and (–) no stop codon were confirmed by restriction enzyme PVUII, and SlSAMT with BbSI and MluI followed by Sanger sequencing using M13 universal primers. All the positive clones were aligned using online MUSCLE software. The positive BP clones were incorporated into binary expression vectors pENSG-YFP (N-terminal) and pEXSG-YFP (C-terminal) using LR Clonase II enzymes (Gateway ™ Technology, Invitrogen). The final LR clones were confirmed by digesting with enzymes AvaI for XSP10 and EcoRV for the SlSAMT gene with ( +) stop codon and (–) no stop codon orientation. Virtual gateway cloning constructs were designed using the Vector NTI software tool (Thermo Fisher, Life Technologies).
Generation of Gateway split-YFP constructs
The target genes were PCR amplified with Gateway adapter primers without stop codon and purified by Mini elute gel extraction kit (Qiagen, Germany) and cloned in pDNR221 by BP Clonase II (Gateway ™ Technology, Invitrogen). The BP clones of both SlTRAX and SlCCoAOMT constructs were confirmed by restriction enzyme PVUII followed by Sanger sequencing using M13 primers. The BP clones were cloned into split-YFP expression vectors pE-SPYNE and pE-SPYCE through site-specific recombination using LR Clonase II (Gateway ™ Technology, Invitrogen). The gateway split-YFP constructs pE-SPYNE directs N-terminals fusion and pE-SPYCE for C-terminals fusion (Gehl et al. 2009). The final destination clones of SlTRAX and SlCCoAOMT were confirmed by digesting with restriction enzymes PVUII and EcoRI.
Tomato protoplast isolation and transformation
The young leaves of 3-week old tomato seedlings were collected with a sterile scalpel for harvesting healthy protoplast following the protocol standardized for Arabidopsis with little modification (Yoo et al. 2007). Briefly, fresh enzymatic solution (1.5% cellulase, 0.4% macerozyme, 20 mM MES at pH 5.7, 0.4 M tris–Hcl, 10 mM CaCl2, 20 mM KCl, 0.1% BSA) was prepared and the tissues digested for 4 h in dark with shaking at 40 rpm. The digested protoplasts were filtered using 75 µm nylon mesh. The filtered protoplasts were centrifuged at 100 g for 2 min and the pellet was resuspended with pre-cooled W5 solution (154 mM NaCl, 125 mM Cacl2, 5 mM KCl, 2 mM MES at pH-5.7, 0.1 M glucose. Then finally resuspended in MMG solution (400 mM mannitol, 15 mM Mgcl2, 4 mM MES at pH 5.7). For protoplast transformation, 2 µg of DNA was gently transfected into 100µL healthy tomato protoplasts with 40% PEG (PEG-4000) solution. Protoplasts transfected with empty plasmids were used as negative controls. The transformed protoplasts were cultured in dark at room temperature overnight and observed under confocal laser scanning microscopy (Leica Microsystems, Germany). Each experiment was repeated three times.
Particle bombardment (Gene delivery) using PDS1000 He system
Bombardment of constructs into tomato seedlings was carried out with little modifications using the PDS1000 He system as described (Ueki et al. 2013). Briefly, ~ 5 µg of DNA of the target construct was coated with 30 mg of 1 µm gold particles. The bombardment was carried out on the abaxial side of the cotyledons of tomato seedlings with a helium pressure of 1100 psi. After bombardment, cotyledons were incubated in dark for 24 h inside a petri dish with wet filter paper and observed under confocal laser scanning microscopy.
Confocal laser scanning microscopy
The transfected tomato protoplast cells and seedlings with respective constructs were visualized in TCS SP5 confocal laser-scanning microscope (Leica Microsystems, Wetzlar, Germany). YFP was excited with the argon laser (488 nm). The fluorescence of YFP was detected using the emission filters, BP500-530 nm, and BP555-615 nm, respectively. A red filter was used as a negative control to detect autofluorescence emitted by chlorophyll pigment (Guadagno et al. 2017). Images were acquired sequentially line-by-line with a resolution of 512 × 512 pixels and 400-Hz scanning speed. Images were processed and exported in TIFF format. The threshold peak of the YFP region of interest (ROI) was quantified by L-XAS in-built confocal software.
Fusarium wilt assay
The pathogenic fungal strain Fusarium oxysporum f.sp. lycopersici Snyder and Hansen (1322) was procured from ITCC, New Delhi. The fungus was grown in PDA for five days at 27°C. Using the root dip method (Mes et al. 1999), tomato seedlings of Arka Vikas (AV) and Arka Abhed (AA) were inoculated with virulent Fol1322. For bioassay, 4-week-old tomato seedlings were placed in spore suspension (0.5 × 107 spores/ml), and inoculated plantlets were immediately re-potted in the soil. A disease progression assessment was performed after three weeks of post-infection. Plant weight and disease index score were examined for 20 plants/treatment. Disease severity was estimated using a Disease Index (DI) by grading 0–4, using the formula [0, no symptoms; (1) slightly swollen or bent hypocotyl; (2) one or two brown vascular bundles in hypocotyl; (3), at least two brown vascular bundles and growth distortion (strong bending of the stem and asymmetric development); (4) all vascular bundles are brown, plant either dead or very small and wilted] (Rep et al. 2004; van der Does et al. 2019). A statistical one-way analysis of variance (ANOVA) and pairwise comparison with Student's t-test for the weight measurements and the non-parametrical Kruskal–Wallis test for the disease index was performed using PRISM 9.0 GraphPad software (Table S3). Each assay was repeated twice.
Rapid microscopy assay
For microscopy assay, 10–12 days old tomato seedlings were treated as described by (van der Does et al. 2019). The roots were gently washed with sterile water to remove the media attached to the tips of the roots system. Clean seedlings were placed in Petri dishes, with the roots spread out on the bottom of the dish and the hypocotyl leaning to the vertical wall, the cotyledons and leaves sticking outside the petri dish. The dish was filled with 25 ml water, to which Fol spores were added to a final concentration of 0.5 × 107 spores/ml. The roots were stained with trypan blue and inspected microscopically 24 h after inoculation (van der Does et al. 2019). The intensity of root colonization by Fol was measured as described by (de Lamo et al. 2018; Prihatna et al. 2018).
Results
In silico analysis of XSP10 and SlSAMT genes of S. lycopersicum
Based on extensive literature mining and reported functional roles as putative susceptible genes for Fol in tomato (Solanum lycopersicum L.), XSP10 and SlSAMT genes have been chosen for the study. The XSP10 gene (gene ID: 101,258,342) with a genomic region of 674 base pairs (bp) encodes a protein with 112 amino acids and a molecular mass of 10 kDa. It consists of two functional exons. Exon 1 with a size of 321 bp and exon 2 of 17 bp and an N-terminal signal peptide of 21 bp (Fig. 1A). A structural domain trypsin α amylase (21-106 bp) with 4-helices folded topology forming right-handed superhelix, ZnF UBR1 (23-99 bp), a zinc finger N recognition site, and BowB (55–106 bp), a Bowman-Birk type proteinase inhibitor (Fig. 1B). From the Sol-Genomics Network database, amino acid sequences were extracted to study the phylogenetic relationship of XSP10 with those of different Solanaceae family. Comparative analysis of multi-sequence alignment showed rich proline and cysteine residues within the conserved domain structure (Fig. 1C). The molecular phylogenetic analysis of XSP10 showed 100% identity to those of Nicotiana tabacum and Nicotiana tomenlosiformis species, 99% identity to Solanum tuberosum and Solanum pennellii, 96% identity to Solanum chilense and Nicotiana attenuata (Fig. 1D).
Schematic representation of gene structure, multiple sequence alignment, and phylogenetic analysis of XSP10 and SlSAMT genes of tomato. A Gene structure of XSP10 which comprised of two exons interspaced by an intron with a gene size of 3491 kb. B domain structure of XSP10 consists of a signal peptide, trypsin α amylase, and Bow B located within the trypsin amylase domain. C Deduced amino acid sequence alignment of XSP10 with other plants of the Solanaceae family by using DNAMAN software. Identical and conserved amino acid residues are shaded in black (100% similarity) and grey (75%) similarity. D Phylogenetic tree generated by Neighbour-Joining method. Bootstrap values were shown as a percent of 1000 replicates. E Gene structure of SlSAMT which comprised of four exons and three introns with a gene size of 2253 bp. F Conserved domain structures have a common methyl transferase domain. G Deduced amino acid sequence of SlSAMT alignment with other members of the Solanaceae family. H Phylogenetic analysis of SlSAMT compared with the Solanaceae family
Salicylic acid methyl transferase (SlSAMT) of Solanum lycopersicum (gene ID: 100,529,139) consists of a genomic region of 2253 bp. It has a CDS of 1086 bp spanning across 4 exons located on chromosome 9, encoding 361 amino acid residues with a molecular mass of 43.32 kDa (Fig. 1E). The predicted functional domain methyl transferase ranges between 38 and 359 bp (Fig. 1F). The amino acid alignments have shown to contain high glutamic acid (E) residues within the diverse Solanaceae family (Fig. 1G). Molecular phylogenetic analysis of SlSAMT revealed 100% identity to those of Datura wrightii and Atropa belladona and 90% identity to that of Nicandra physalodes (Fig. 1H).
Tissue-specific expression analysis of XSP10 and SlSAMT in different tomato cultivars
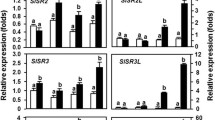
To understand the primary mRNA transcript expression levels of XSP10 and SlSAMT genes in different tomato cultivars, semi-quantitative PCR (RT-PCR) and qRT-PCR were performed. RT-PCR was performed initially to test the expression pattern in different tissues. It was observed from the RT-PCR results that XSP10 showed its expression in all the tissues across three cultivars tested while strong expression was observed in root and least in leaf (Fig. 2A–C). The fruit tissue with the least expression was taken as a control for measurement of relative expression (Fig. 2D).
Expression analysis of XSP10 and SlSAMT genes in different tissues of three different tomato cultivars through semi-quantitative PCR and qRT-PCR. A–C Semi-quantitative PCR based expression of XSP10 and SlSAMT in different tissues (root, stem, leaf, flower, fruit and seed) of tomato cultivars MT, AV, and AA. The expected amplicon size was 118-bp for EF1α, 114-bp for XSP10, and 120-bp for SlSAMT. EF1α was used as an internal control. Expression quantification of XSP10 (D) and SlSAMT (E) genes in different tissues of three tomato cultivars. EF1α was used as a house-keeping gene control. All statistical data were calculated in mean ± standard deviation. The error bar represents the standard deviation of the sample. The level of significance was performed by parametric t-test (*p < 0.05), **p < 0.01, ***p < 0.001) using XLSTAT software. The asterisk symbol indicates significant differences. The experiment was set up with three biological replicates and three technical replicates for each sample. Each experiment was repeated thrice and the results were concordant
The expression of XSP10 in different tissues of three different tomato cultivars was further quantified and analyzed by qRT-PCR. MT showed a highly upregulated expression of XSP10 in the root (13.8 fold) followed by the stem (4.5 fold), flower (4.0 fold), leaf (0.2 fold), and seed (0.2 fold). AV showed the highest expression of XSP10 in roots (10.17 fold) followed by the stem (9.8 fold), leaf (6.0 fold), flower (5.1 fold), and seed (2.6 fold) (Fig. 2D). AA also showed the highest expression of XSP10 in the root (8.7 fold), followed by the stem (6.6 fold), flower (3.1 fold), leaf (2.6 fold), and seed (0.7 fold) (Fig. 2D). The results indicate higher expression of the XSP10 gene in roots in general in all three cultivars tested. Interestingly, XSP10 showed higher expression in AV compared to AA in most of the tissues tested (Fig. 2D).
Similarly, the expression level of SlSAMT in different tissues of three tomato cultivars was measured. It was observed that the SlSAMT gene showed expression in all the tissues across three cultivars tested while strong expression was observed in flowers and least in fruit and seed (Fig. 2A–C). Through qRT-PCR it was observed that in MT, the highest expression level of SlSAMT was observed in flower (8.9 fold) followed by leaf (5.3 fold), root (2.6 fold), stem (1.9 fold), and fruit (1.9 fold) while seed was chosen as a basal reference for qRT-PCR because of its least expression (Fig. 2E). In AV, the highest expression of SlSAMT was observed in flower (9.3 fold) followed by the stem (8.6 fold), leaf (5.6 fold), root (3.1 fold) fruit (0.7 fold) (Fig. 2E). AA showed mRNA expression levels of SlSAMT highest in flower (7.6 fold), followed by leaf (5.2 fold), stem (5.1 fold), fruit (1.6 fold), and root (1.1 fold). Interestingly, SlSAMT showed highly up-regulated expression in wilt susceptible cultivar (AV) compared to multiple disease resistant cultivars (AA) in most of the tissues tested (Fig. 2E).
Overall, the results showed that XSP10 strongly expressed in root while SlSAMT in flower tissue in all three tomato cultivars while both genes showed highly upregulated expression in wilt susceptible tomato cultivar (AV) than multiple disease resistant cultivar (AA). The qRT-PCR data with detailed statistical analysis of XSP10 and SlSAMT genes is given in Table S2.
Analysis of sub-cellular localization of XSP10 and SlSAMT
Despite the importance of XSP10 and SlSAMT as susceptible genes to Fol, little is known about their cell biology. The current study aimed to understand their sub-cellular localization through the transient cell biological approach in protoplasts and seedlings of tomato (S. lycopersicum). XSP10 was predicted to be localized to the cell membrane, with a maximum score of 4.27, and nucleus (0.198) and SlSAMT predicted to be localized to the cytoplasm (2.44) and nucleus (1.12) (Fig. S1 A–B). To study their sub-cellular localization in vivo, XSP10 and SlSAMT have been cloned into binary plant expression vectors with N and C terminal YFP fusions using Gateway cloning technology. The virtual gateway cloning information of subcellular localization constructs is given in Fig. S2. The positive clones were further confirmed by Sanger sequencing (Fig. S3 A–D) and restriction digestion (Fig. S1). YFP:XSP10 and XSP10:YFP showed predominant localization at the nucleus as well as at the cell membrane in tomato protoplasts (Fig. 3A, C). The empty expression vectors pENSG-YFP and pEXSG-YFP used as negative controls did not show any YFP signals (Fig. 3B, D). As an independent confirmation, YFP:XSP10 and XSP10:YFP constructs were transformed into tomato seedlings through gene gun PDS 1000/He system and observed through confocal microscopy. Both YFP: XSP10 and XSP10:YFP showed predominant localization at the nucleus (Fig. 3E, F). The localization signals of XSP10: YFP were clear and consistent compared to YFP: XSP10.
Sub-cellular localization of N and C terminal YFP fusions of XSP10 in tomato protoplasts and seedlings under confocal microscopy. Tomato protoplasts transformed with N terminal (A) and C terminal (C) YFP fusion of XSP10 which showed its predominant localization in the nucleus. Tomato seedlings transformed through the PDS/1000 system with N terminal (E) and C terminal (F) fusion of XSP10 also showed strong localization in the nucleus. DAPI stained cells of each construct were used as a comparative marker that showed a clear nuclear signal (a, e, i, m, q, u). Tomato protoplasts transformed with empty plasmid pENSG-YFP used a negative control for N terminal (B) and pEXSG-YFP for C terminal (D) YFP fusions which did not show any YFP signal (f and n). Arrows highlight the sub-cellular localization of XSP10 in the nucleus and DAPI stained cells in the nucleus only. Scale bar 25 μM
Tomato protoplast cells transformed with YFP: SlSAMT and SlSAMT: YFP showed strong in vivo sub-cellular localization in the cytoplasm (Fig. 4A, C). The negative controls pENSG-YFP and pEXSG-YFP did not show any YFP signals (Fig. 4B, D). Tomato seedlings independently transformed with YFP: SlSAMT and SlSAMT: YFP through gene gun PDS 1000/He system also showed clear in vivo localization in the cytoplasm (Fig. 4E, F). To exclude the false positives/negatives, the fluorescent signals of each construct of XSP10 and SlSAMT were quantified using XLS confocal software in comparison to negative controls and found to be true fluorescent signals (Fig. S4 A–J). A comprehensive summary of in silico and in vivo sub-cellular localization of XSP10 and SlSAMT along with available literature to-date is given in Table 1.
Sub-cellular localization of N and C terminal YFP fusions of SlSAMT in tomato protoplasts and seedlings under confocal microscopy. Tomato protoplasts transformed with N terminal (A) and C terminal (C) YFP fusion of SlSAMT showed clear localization in the cytoplasm. Tomato seedlings transformed through the PDS/1000 system with N terminal (E) and C terminal (F) fusion of SlSAMT also showed strong localization in the cytoplasm. DAPI stained cells of each construct which was used as a comparative marker show a clear nuclear signal (a, e, i, m, q,u). Tomato protoplasts transformed with empty plasmid pENSG-YFP used as a negative control for N terminal (B) and pEXSG-YFP as C terminal (D) YFP fusions which did not show any YFP signal (f and n). Arrows highlight the sub-cellular localization of SlSAMT in the cytoplasm while DAPI stained cells in the nucleus. Scale bar 25 μM
In vivo protein–protein interaction of XSP10 and SlSAMT with stress-responsive proteins
Fusarium wilt is a complex ubiquitous disease that might be regulated by multiple host genes and there may be a cross-talk of plant-microbial interaction and plant defense pathway genes involved. Hence it is necessary to identify the interacting partners as regulatory proteins that might be functioning together in imparting tolerance to Fusarium wilt. Accordingly, the in silico network data prediction showed a strong interaction between XSP10 & SlTRAX with a score of 0.81; strong interaction between SlSAMT & SlCCoAOMT with a score of 0.771 and XSP10 and SlSAMT with a score of 0.41. The nodes describe the protein encoding by a single gene and edges for the association of specific binding of proteins (Fig. S5 A–C).
To validate the in silico findings and identify the protein interactors of XSP10 and SlSAMT in vivo, a systematic Split-YFP (Bi-FC) assay was performed in tomato protoplasts. Here the split-YFP constructs of XSP10 and SlSAMT and putative interacting predicted partners SlCCoAOMT and SlTRAX with no stop codon were tagged with either the N- or C-terminal half of yellow fluorescent protein (YFP) variants. The split-YFP constructs of each partner were generated in split-YFP expression vectors pE-SPYNE and pE-SPYNE using the Gateway cloning approach (Fig. S5 d–k). The virtual split-YFP constructs were generated initially using the Vector NTI software tool (Thermo Fisher, Life Technologies) (Fig. S1). The split-YFP constructs were transformed into tomato protoplasts and visualized under confocal microscopy. XSP10 showed strong interaction with SlSAMT and the interaction was observed to be taking place at the cytoplasm (Fig. 5A). XSP10 also showed strong interaction with SlTRAX and the site of interaction was in the cytoplasm (Fig. 5B). SlSAMT showed moderate interaction with SlCCoAOMT in the cytoplasm (Fig. 5C). A very weak interaction was also observed between SlSAMT and SlTRAX (Fig. 5D) as well as between XSP10 and SlCCoAOMT (Fig. 5E). Empty split-YFP expression vectors co-expressed in protoplasts were used as negative controls which did not show any YFP signals (Fig. 5F). The strength of the interaction between the proteins was determined based on the fluorescence quantification signals measured by XLS confocal software as represented in (Fig. S4 k–p). A comprehensive comparison summary of in silico and in vivo interactions of XSP10, SlSAMT, and stress-responsive proteins SlTRAX and SlCCoAOMT are given in Table 1.
In vivo protein–protein interaction of XSP10, SlSAMT, and stress associated proteins in tomato protoplasts under confocal microscopy. A. Tomato protoplasts co-transformed with split-YFP constructs of SlSAMT and XSP10 proteins showed clear signals in the cytoplasm. B Strong positive interaction of XSP10 with SlTRAX at cytoplasm. C Moderate interaction of SlSAMT with SlCCOAOMT at cytoplasm. D Weak interaction of SlSAMT with SlTRAX. E Weak interaction of XSP10 with SlCCOAOMT. F Tomato protoplasts co-transformed with empty plasmid used as a negative control for split-YFP fusions which did not show any YFP signal. DAPI stained cells of each construct which was used as a comparative marker show a clear nuclear signal (a, e, i, m, q, u). Arrows highlight the strong interactions (b, f, j) while DAPI stained cells in the nucleus. Scale bar 8 μm, 10 μm and 25 μm
Phenotypic evaluation of wilt disease in cultivars Arka Vikas and Arka Abhed to Fol 1322 infection
To study the role of fungal root colonization, a rapid microscopy assay was performed in 10–12 days old seedlings of wilt susceptible cultivar AV and multiple disease resistant cultivar AA. Atleast 20 roots of each cultivar were examined under microscopy. Our result shows early stages of susceptible wilt root colonization in AV compared to AA (Fig. 6 A, B). Microscopy observation reveals < 84% of fungal mycelium colonizes in root hairs of AV while below 25% of fungal hyphae colonized in AA. After 24 h, hyphae were found to attached the root hairs and colonized more intensively at the root surface and apex. After attachment, hyphae started to grow predominantly along the junction of the epidermal cells. In addition, there was a mass hyphal growth of Fol in root hairs of AV compared to AA. Moreover, the significance difference (p*** < 0.001) was evaluated and the intensity of root colonization was higher in AV compared to AA (Table S3).
Phenotypic evaluation of wilt susceptible cultivar Arka Vikas (AV) and multiple disease resistant cultivar Arka Abedh (AA) to Fol 1322 infection. Root colonization by Fol at 24 h after inoculation observed using bright-field microscopy (A). The red arrow indicates fungal colonization to the root surface. The intensity of colonization was measured as the area of mycelia against the total area of an image and quantified by Image J software (B). Image quantification was based on gray levels of 8-bit images. The asterisk symbol represents a significant difference (***P < 0.001). Root dip bioassay of 4 –weeks old tomato seedlings of Arka Vikas and Arka Abhed (C) inoculated with Fol 1322 observed at 21 days post inoculation. (D) Disease symptoms were scored by measuring the fresh weight above the cotyledon and (E) disease index (0–4) of independent 20 plants/treatment. Plant weight was subjected to a pairwise comparison Students t-test, whereas disease index (DI) was determined by non-parametric Kruskal–Wallis test (*p < 0.05,**p < 0.01,***p < 0.001) using PRISM.9.0 GraphPad software
Also, to test the response of wilt disease phenotype in susceptible cultivar AV and multiple diseases resistant cultivar AA. The Fol inoculation bioassay (Mes et al. 1999) was carried out on four-week-old plants of AV and AA inoculated with Fol 1322 to define the negative regulatory function and susceptibility nature of XSP10 and SlSAMT genes. After three weeks of post-inoculation, average fresh weight and the disease index were calculated in both the tomato cultivars. Stunting, chlorosis, necrosis, vascular wilt and cell death are the disease symptoms of the plant caused by Fol (Di et al. 2017; Prihatna et al. 2018). Phenotypic data analysis revealed that AV leaves infected with Fol showed chlorotic, leaf epinasty and mild wilt symptoms than AA, as shown in Fig. 6C. The disease index (DI) of 20 plants/ treatment was measured to each cultivar upon Fol1322 infection. The average weight of the infected plants was calculated. The fresh weight of infected AA is significantly higher (p*** < 0.001) compared to infected AV (Fig. 6D). The DI of AA plants was significantly (p** < 0.01) attenuated relative to that of AV plants (Fig. 6E). The result was consistent in all two repetitions (Table S3). Overall, our data suggested that fusarium wilt disease symptoms were higher in AV than AA upon Fol1322 infection.
Discussion
XSP10 is predominantly expressed in root and SlSAMT is highly expressed in flower tissue of tomato cultivars
XSP10 is a lipid binding protein of S. lycopersicum whose tissue-specific expression was not known. The present study revealed the strong expression of XSP10 in the root tissue of different tomato cultivars. XSP30, a xylem sap protein of Cucumis sativus was shown to specifically express in roots but could not detect in any other organs (Masuda et al. 1999). The highly upregulated expression of XSP10 in root tissue of tomato cultivars indicates its possible regulatory function in plant defense during pathogen penetration from root tissues.
The present study revealed the predominant expression of SlSAMT in flower tissues of different tomato cultivars. It was reported earlier that SlSAMT was highly expressed in flower buds with low expression in young and mature leaves and no expression in other tissues (Tieman et al. 2010). Our data of expression of SlSAMT is consistent with the work (Tieman et al. 2010). The higher expression of SlSAMT in flowers might be due to its involvement in catalytic properties in tissue-specific protection and volatile molecule formation (Tieman et al. 2010). It was demonstrated that AtBSMT1 can be dispersed in sepals of flowers, leaf trichomes, and hydathodes under normal growth conditions in Arabidopsis (Chen et al. 2003). Transcriptional and post-translational regulation of SAMT has shown methyl transferase involved in flower development of Stephanotis floribunda (Pott et al. 2003). In our study, SlSAMT transcripts level was higher in AV compared to MT followed by AA with the least mRNA expressed in the fruit. This indicates a low MeSA accumulation by the catalytic activity of SlSAMT-encoding enzymes in the fruit. It confirms earlier speculation that SlSAMT activity may decline in fruits due to early biosynthesis of MeSA in early developing organs (Tieman et al. 2010). In agreement with our previous studies, MeSA, a derivative of salicylate might be involved in early biological processes during fruit ripening by modulating the SA biosynthetic pathways.
There are three physiological races of pathogen isolates (1, 2, and 3) that are distinguished by their host-specific pathogenicity in tomato cultivars (Nirmaladevi et al. 2016). XSP10 was tested with F. oxysporum f. sp. lycopersici (Fol) isolate either with a virulent race 2 isolates of Fol (Fol007) or with an avirulent race 1 isolate Fol004 (Rep et al. 2005) in tomato (Solanum lycopersicon cv. Moneymaker GCR161). It was reported that XSP10-silenced lines did not affect I-mediated resistance and it remained fully resistant against the avirulent Avr1-carrying Fol004 isolate race1 (Krasikov et al. 2011). However, seedlings inoculated with the virulent strain Fol007 revealed a significantly high average weight, a lower disease index, and a smaller percentage of dead plants after infection in the XSP10-silenced plants (Krasikov et al. 2011). Similarly, the previous data also revealed that the SlSAMT gene was found disease susceptibility in tomato cultivar Moneymaker GCR161 inoculated with race-2 isolate of F. oxysporum f. sp. lycopersici (Fol007; virulent on GCR161) (Ament et al. 2010). However, silencing of SlSAMT did not affect I-mediated resistance of GCR161 to the race-1 isolate Fol004 (Ament et al. 2010).
The non-uniformity of expression of XSP10 and SlSAMT in different tomato cultivars could be due to the genotypic variations between the cultivars as expected. The variations could also be due to natural exposure of organs associated with various stresses induced by symbiotic pathogens (Liu et al. 2012). At the molecular level, the reason for the variation of gene expression might be due to the involvement of gene conformity and cis-acting promoter structure (Das and Bansal 2019).
Interestingly, both XSP10 and SlSAMT genes have shown highly upregulated expression in AV, a known wilt susceptible cultivar (Upreti and Thomas 2015) than AA, a known multiple disease resistant tomato cultivar (Nirmaladevi et al. 2016). The results also support by the fact that the TDF (transcript-derived fragment) gene expressed highly in a susceptible cultivar of Hop than a resistant cultivar upon Verticillium albo-atrum infection (Cregeen et al. 2015). These findings suggest that both XSP10 and SlSAMT as Fusarium wilt disease susceptible genes might likely co-expressed and negatively regulate genetic tolerance to Fol.
XSP10 is predominantly localized to the nucleus and SlSAMT is localized to the cytoplasm
The computational approach of predicting subcellular localization of proteins has been an area of interest in recent years (Dönnes and Höglund 2004). CELLO2GO software is a reliable and user-friendly tool to predict protein localization (Yu et al. 2014). By targeting the query sequences, the in silico localization of XSP10 was predicted in the extracellular region and the nucleus. On the other hand, transient studies are an important and reliable alternative to stable plant transformation due to the cost-effective and non-tedious nature of techniques (Huo et al. 2017; Baruah et al. 2020; Saikia et al. 2020).
Few xylem sap proteins of Brassica oleraceae were found to be located in the cell wall (Ligat et al. 2011). These small proteins might involve in the transfer of lipid molecules to the extracellular space and accompany few other ns-LTPs proteins of the prolamine superfamily for recycling in the endosperm, and seed wall development (Eklund and Edqvist 2003). It was shown that the majority of non-specific lipid transfer protein (nsLTPs) localized to apoplastic space (Salminen et al. 2016). Over 54% of the identified XSPs have been predicted to be localized in intracellular space, which might be present in exosomes of tomato (de Lamo et al. 2018). In Gossypium sp, XSPs have a significant contribution to the reinforcement of cell wall (Zhang et al. 2015).
In silico analysis of the current study showed the localization of XSP10 to cell membrane and nucleus while the in vivo analysis in both protoplasts and seedlings of tomato showed its predominant localization in the nucleus. Our findings suggest that the XSP10 protein at the nuclear compartment may play a significant role not only in the post-translation and protein modifications but also in transcriptional activation during stress response function (Salminen et al. 2016). It might also be involved in lipid molecule signaling during pathogenic attacks (Salminen et al. 2016) and cell wall development (Zhang et al. 2015).
Salicylic acid (SA) is an important plant hormone which activates and regulates metabolic pathway in response to abiotic and biotic stresses (Hayat et al. 2010). As a mobile signal, SA is first detected in the cytoplasm, later in the chloroplast, and stored in the vacuole in the glycosylated form (Maruri-López et al. 2019). CELLO2GO predicted the localization of SlSAMT protein mainly to the cytoplasm. The in vivo study in protoplast and seedlings of tomato also showed its predominant localization to the cytoplasm. A previous study in snapdragon flower showed localization of SAMT in the cytosol (Kolosova et al. 2001). This cytosolic protein may regulate cellular and metabolic signaling pathways apart from the indirect and direct response during fungal-host tomato interaction (Ament et al. 2010). The current findings also reveal that many uncharacterized novel stress derived cytosolic proteins may modulate SlSAMT proteins with a functional cross-talk of any hormonal signaling pathways but prone to Fusarium stress susceptibility (FSS). Taken together, the in vivo sub-cellular localization results revealed the site of the function of XSP10 as the nucleus and SlSAMT as the cytoplasm.
XSP10 showed strong interactions with SlSAMT and SlTRAX in the cytoplasm
The biological interactomics system is evaluated by gene ontology (GO), domain-motif interaction (DMI), co-expression, and co-localization (Yue et al. 2016). The in silico approach has helped to integrate systematic annotation of different pipelines of in vivo protein–protein interactions (PPIs) of XSP10, SlSAMT, and their stress-responsive partners (Fig.S4 a–c).
Split-YFP is a well-established method for transient in vivo PPIs in plant cells (Gehl et al., 2009). From the current study, a strong interaction of XSP10 was observed with SlSAMT in the cytoplasm. XSP10 localized to the nucleus and cell membrane while SlSAMT localized to cytoplasm however, their in vivo interaction taking place at the cytoplasm. This suggests that XSP10 is not only transcriptionally activated but trans-located later in presence of SlSAMT in the cytoplasm to carry out stress response function (Dempsey et al. 2011). This result shows the functional accountability of post-translational modification. The strong interaction between XSP10 and SlSAMT unveils the cellular and molecular connectivity, which might influence a functional role during Fol colonization. SlTRAX is a DNA binding protein having both I-2 CC and NB-ARC domains. TRAX, a stress-associated protein is known to be localized in the cytoplasm of Arabidopsis thaliana and Oryza sativa (Chennathukuzhi et al. 2001; Chittela et al. 2014). The strong interaction at cytoplasm suggests that nuclear localized proteins XSP10 and SlTRAX might shuttle from the nucleus to cytoplasm and involved in a fundamental role in vesicular trafficking, mRNA transport, and signal transduction. An earlier report showed TRAX was involved in translational processes, mRNAs vesicular trafficking, and nuclear localization signal peptide in mammalian cells (Chennathukuzhi et al. 2001). During Fol colonization, SlTRAX might play a negative role along with XSP10. SlSAMT showed strong interaction with CCoAOMT in the cytoplasm. CCoAOMT is an enzyme that falls under O-methyl transferase family and composed of monolignol (Walker et al. 2016). They are mainly active in the cytosol of monocots i.e. sugarcane and maize (Ruelland et al. 2003). In Zea mays, ZmCCoAOMT2 is not only associated with lignin and secondary cell wall formation but also confer resistance to multiple pathogen attacks (Yang et al. 2017). This strong evidence gives a clue where SlSAMT protein might function as a negative regulator of Fol suppressing the positive functions of SlCCoAOMT. This is also evident from the indirect (weak) interaction of XSP10 with SlCCoAOMT as well as SlSAMT with SlTRAX.
Often, split-YFP fluorescence might give false positive signals or artifacts. However, the systematic fluorescence quantification of each interacting partner in comparison to negative controls has excluded this possibility. In vivo results were in agreement with in silico interaction predictions. Taken together, in vivo protein–protein interaction results suggest that XSP10, SlSAMT, and SlTRAX function and co-expressed together in regulating responses to Fusarium stress susceptibility (FSS) in tomato.
XSP10 and SlSAMT enhance wilt disease susceptibility in cultivar Arka Vikas compared to Arka Abhed upon Fol 1322 infection
The pathogen F.oxysporum lycopersici (Fol) develops the disease symptoms by infecting the roots of tomato plants (Prihatna et al. 2018). The consistent difference was correlated in root colonization between rmc and 76 R by Fol Race 3 isolates. Thus, it provides a reliable assessment to differ the phenotypes to Fol Race 3 tolerance and susceptibility (Prihatna et al. 2018). It has been reported that root hairs are the entry point for fungal hyphae into the upper and inner surface of maize. It could happen as the roots of resistant maize lines have few root hairs and are less heavily colonized than susceptible lines (Wu et al. 2013). So far, several aspects of this study exemplified the epidermal cell recognition, colonization and root infection by gfp-labeled F.oxysporum f.sp. radicis lycopersici in tomato (Lagopodi et al. 2002). In case of wilt disease caused by Fol, the root tips are among the other sites for initial infection (Olivain and Alabouvette 1999). The genetic diversity and pathogenicity virulence caused by Fol strains were observed in tomato cultivar AV. The pathogenic strains resulting in vascular wilt disease in different tomato cultivars solely depend on their virulence (Nirmaladevi et al. 2016). Besides contributing to virulence, few effector molecules were recognized by the tomato resistance gene (R-genes) products conferred avirulence to a fungal pathogen (Cao et al. 2018). Differential expression of genes upon infection with Verticillium albo-atrum has indicated higher fungal infection in susceptible Hop cultivar than resistant Hop cultivar (Cregeen et al. 2015). It is generally understood that the resistance trait is under the molecular mechanism of genetic control (Upreti and Thomas 2015). The concept of genetic variation in susceptible cv. AV and multi-resistant cv. AA indicates XSP10 and SlSAMT might play a negative role in genetic tolerance to Fol. qRT-PCR results of our study are in accordance with these findings where XSP10 and SlSAMT were highly expressed in AV than AA. Taken together, our findings suggest that XSP10 and SlSAMT enhance fusarium wilt disease susceptibility in cv. AV compared to multiple disease resistant cv. AA upon Fol infection.
Conclusions and future prospects
The present study revealed the tissue-specific expression of XSP10 and SlSAMT, two putative disease susceptible genes associated with Fusarium wilt of tomato. The highly up-regulated expression of these two genes in biotic stress susceptible tomato cultivar than a resistant cultivar hints at their possible role as negative regulators of Fusarium wilt tolerance. XSP10 localized predominantly at the nucleus while SlSAMT localized to the cytoplasm. However, they both interact at cytoplasm in vivo along with SlTRAX. A strong in vivo interaction of XSP10 with SlSAMT and stress-associated SlTRAX in the cytoplasm indicates their possible functional site for Fusarium wilt tolerance response is cytoplasm.
The current study is highly significant in providing cell and molecular insights for understanding the negative regulatory mechanism of XSP10 and SlSAMT in Fusarium wilt tolerance. A systematic genetic analysis of XSP10 and SlSAMT through dual-gene CRISPR/Cas editing and functional analysis in stably transformed tomato lines would reveal their role in genetic tolerance to Fol.
Accession numbers
Sequence data from this article can be found in Gene Bank/EMBL data libraries under accession number XSP10 (NC_015440), SlSAMT (NC_015446), SlTRAX (NC_015441) and SlCCoAOMT (NP_001234801).
Abbreviations
- XSP:
-
Xylem sap protein
- SAMT:
-
Salicylic acid methyl transferase
- MeSA:
-
Methyl salicylate
- RT-PCR:
-
Reverse transcriptase polymerase chain reaction
- qRT-PCR:
-
Quantitative real time polymerase chain reaction
- PPI:
-
Protein–protein interactions
- Split-YFP:
-
Split-yellow fluorescence protein
- Bi-FC:
-
Bimolecular fluorescence complementation
- TRAX:
-
Translin-associated factor X
- CCoAOMT:
-
Caffeoyl-CoA O-methyltransferase
References
Alvarez S, Marsh EL, Schroeder SG, Schachtman DP (2008) Metabolomic and proteomic changes in the xylem sap of maize under drought. Plant Cell Environ 31:325–340. https://doi.org/10.1111/j.1365-3040.2007.01770.x
Ament K, Krasikov V, Allmann S et al (2010) Methyl salicylate production in tomato affects biotic interactions. Plant J 62:124–134. https://doi.org/10.1111/j.1365-313X.2010.04132.x
Baruah I, Baruah G, Sahu J et al (2020) Transient sub-cellular localization and in vivo protein-protein interaction study of multiple abiotic stress-responsive AteIF4A-III and AtALY4 proteins in Arabidopsis thaliana. Plant Mol Biol Report 38:538–553. https://doi.org/10.1007/s11105-020-01219-w
Biles CL, Abeles FB (1991) Xylem sap proteins. Plant Physiol 96:597–601. https://doi.org/10.1104/pp.96.2.597
Blein JP, Coutos-Thévenot P, Marion D, Ponchet M (2002) From elicitins to lipid-transfer proteins: a new insight in cell signalling involved in plant defence mechanisms. Trends Plant Sci 7:293–296. https://doi.org/10.1016/S1360-1385(02)02284-7
Buhtz A, Kolasa A, Arlt K et al (2004) Xylem sap protein composition is conserved among different plant species. Planta 219:610–618. https://doi.org/10.1007/s00425-004-1259-9
Cao L, Blekemolen MC, Tintor N et al (2018) The Fusarium oxysporum Avr2-Six5 effector pair alters plasmodesmatal exclusion selectivity to facilitate cell-to-cell movement of Avr2. Mol Plant 11:691–705. https://doi.org/10.1016/j.molp.2018.02.011
Chen F, D’Auria JC, Tholl D et al (2003) An Arabidopsis thaliana gene for methylsalicylate biosynthesis, identified by a biochemical genomics approach, has a role in defense. Plant J 36:577–588. https://doi.org/10.1046/j.1365-313X.2003.01902.x
Chennathukuzhi VM, Kurihara Y, Bray JD, Hecht NB (2001) Trax (Translin-associated Factor X), a Primarily Cytoplasmic Protein, Inhibits the Binding of TB-RBP (Translin) to RNA. J Biol Chem 276:13256–13263. https://doi.org/10.1074/jbc.M009707200
Chittela RK, Gupta GD, Ballal A (2014) Characterization of a plant (rice) translin and its comparative analysis with human translin. Planta 240:357–368. https://doi.org/10.1007/s00425-014-2092-4
Cregeen S, Radisek S, Mandelc S et al (2015) Different gene expressions of resistant and susceptible Hop cultivars in response to infection with a highly aggressive strain of Verticillium albo-atrum. Plant Mol Biol Report 33:689–704. https://doi.org/10.1007/s11105-014-0767-4
Das S, Bansal M (2019) Variation of gene expression in plants is influenced by gene architecture and structural properties of promoters. PLoS ONE 14:e0212678. https://doi.org/10.1371/journal.pone.0212678
de Lamo FJ, Constantin ME, Fresno DH et al (2018) Xylem Sap proteomics reveals distinct differences between R gene- and endophyte-mediated resistance against Fusarium Wilt disease in tomato. Front Microbiol 9:1–13. https://doi.org/10.3389/fmicb.2018.02977
Dempsey DA, Vlot AC, Wildermuth MC, Klessig DF (2011) Salicylic acid biosynthesis and metabolism. The Arabidopsis Book 9:e0156. https://doi.org/10.1199/tab.0156
Di X, Gomila J, Takken FLW (2017) Involvement of salicylic acid, ethylene and jasmonic acid signalling pathways in the susceptibility of tomato to Fusarium oxysporum. Mol Plant Pathol 18:1024–1035. https://doi.org/10.1111/mpp.12559
Dönnes P, Höglund A (2004) Predicting protein subcellular localization: past, present, and future. Genomics Proteomics Bioinformatics 2:209–215. https://doi.org/10.1016/S1672-0229(04)02027-3
Dresselhaus T, Hückelhoven R (2018) Biotic and abiotic stress responses in crop plants. Agronomy 8:8–13. https://doi.org/10.3390/agronomy8110267
Effmert U, Saschenbrecker S, Ross J et al (2005) Floral benzenoid carboxyl methyltransferases: From in vitro to in planta function. Phytochemistry 66:1211–1230. https://doi.org/10.1016/j.phytochem.2005.03.031
Eklund DM, Edqvist J (2003) Localization of nonspecific lipid transfer proteins correlate with programmed cell death responses during endosperm degradation in Euphorbia lagascae seedlings. Plant Physiol 132:1249–1259. https://doi.org/10.1104/pp.103.020875
Gawehns F, Ma L, Bruning O et al (2015) The effector repertoire of Fusarium oxysporum determines the tomato xylem proteome composition following infection. Front Plant Sci 6:1–17. https://doi.org/10.3389/fpls.2015.00967
Gehl C, Waadt R, Kudla J et al (2009) New GATEWAY vectors for high throughput analyses of protein-protein interactions by bimolecular fluorescence complementation. Mol Plant 2:1051–1058. https://doi.org/10.1093/mp/ssp040
Goswami RS, Kistler HC (2004) Heading for disaster: Fusarium graminearum on cereal crops. Mol Plant Pathol 5:515–525. https://doi.org/10.1111/J.1364-3703.2004.00252.X
Guadagno CR, Ewers BE, Speckman HN et al (2017) Dead or alive? using membrane failure and chlorophyll a fluorescence to predict plant mortality from drought. Plant Physiol 175:223–234. https://doi.org/10.1104/pp.16.00581
Hayat Q, Hayat S, Irfan M, Ahmad A (2010) Effect of exogenous salicylic acid under changing environment: a review. Environ Exp Bot 68:14–25. https://doi.org/10.1016/j.envexpbot.2009.08.005
Houterman PM, Speijer D, Dekker HL et al (2007) The mixed xylem sap proteome of Fusarium oxysporum-infected tomato plants. Mol Plant Pathol 8:215–221. https://doi.org/10.1111/j.1364-3703.2007.00384.x
Huo A, Chen Z, Wang P et al (2017) Establishment of transient gene expression systems in protoplasts from Liriodendron hybrid mesophyll cells. PLOSE ONE 1–14. https://doi.org/10.1371/journal.pone.0172475
Joshi R (2018) A review of Fusarium oxysporum on its plant interaction and industrial use. J Med Plants Stud 6:112–115. https://doi.org/10.22271/plants.2018.v6.i3b.07
Kolosova N, Sherman D, Karlson D, Dudareva N (2001) Cellular and subcellular localization of S-Adenosyl-L-Methionine: benzoic acid carboxyl methyltransferase, the enzyme responsible for biosynthesis of the volatile Ester Methylbenzoate in Snapdragon Flowers 1. Plant Physiol 126:956–964
Koo YJ, Kim MA, Kim EH et al (2007) Overexpression of salicylic acid carboxyl methyltransferase reduces salicylic acid-mediated pathogen resistance in Arabidopsis thaliana. Plant Mol Biol 64:1–15. https://doi.org/10.1007/s11103-006-9123-x
Krasikov V, Dekker HL, Rep M, Takken FLW (2011) The tomato xylem sap protein XSP10 is required for full susceptibility to Fusarium wilt disease. J Exp Bot 62:963–973. https://doi.org/10.1093/jxb/erq327
Lagopodi AL, Ram AFJ, Lamers GEM et al (2002) Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici Revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol Plant Microbe Interact 15:172–179. https://doi.org/10.1094/MPMI.2002.15.2.172
Ligat L, Lauber E, Albenne C et al (2011) Analysis of the xylem sap proteome of Brassica oleracea reveals a high content in secreted proteins. Proteomics 11:1798–1813. https://doi.org/10.1002/pmic.201000781
Lima Silva CC de, Shimo HM, de Felício R et al (2019) Structure-function relationship of a citrus salicylate methylesterase and role of salicylic acid in citrus canker resistance. Sci Rep 9:3901. https://doi.org/10.1038/s41598-019-40552-3
Liu PP, Yang Y, Pichersky E, Klessig DF (2010) Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol Plant Microbe Interact 23:82–90. https://doi.org/10.1094/MPMI-23-1-0082
Liu PP, von Dahl CC, Park SW, Klessig DF (2011) Interconnection between methyl salicylate and lipid-based long-distance signaling during the development of systemic acquired resistance in Arabidopsis and Tobacco. Plant Physiol 155:1762–1768. https://doi.org/10.1104/pp.110.171694
Liu Z, Zhao J, Li Y et al (2012) Non-uniform distribution pattern for differentially expressed genes of transgenic rice Huahui 1 at different developmental stages and environments. PLoS ONE 7:1–8. https://doi.org/10.1371/journal.pone.0037078
Liu W, Xie Y, Ma J et al (2015) IBS: An illustrator for the presentation and visualization of biological sequences. Bioinformatics 31:3359–3361. https://doi.org/10.1093/bioinformatics/btv362
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Luo J-S, Zhang Z (2019) Proteomic changes in the xylem sap of Brassica napus under cadmium stress and functional validation. BMC Plant Biol 19:280. https://doi.org/10.1186/s12870-019-1895-7
Maruri-López I, Aviles-Baltazar NY, Buchala A, Serrano M (2019) Intra and extracellular journey of the phytohormone salicylic acid. Front Plant Sci 10:1–11. https://doi.org/10.3389/fpls.2019.00423
Masahito Shikata and Hiroshi Ezura (2016) Plant Signal Transduction. Springer, New York, New York, NY
Masuda S, Sakuta C, Satoh S (1999) cDNA cloning of a novel Lectin-Like Xylem Sap Protein and its root-specific expression in cucumber. Plant Cell Physiol 40:1177–1181. https://doi.org/10.1093/oxfordjournals.pcp.a029504
Mes JJ, Weststeijn EA, Herlaar F et al (1999) Biological and molecular characterization of Fusarium oxysporum f. sp. lycopersici divides race 1 isolates into separate virulence groups. Phytopathology 89:156–160. https://doi.org/10.1094/PHYTO.1999.89.2.156
Negre F, Kolosova N, Knoll J et al (2002) Novel S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme responsible for biosynthesis of methyl salicylate and methyl benzoate, is not involved in floral scent production in snapdragon flowers. Arch Biochem Biophys 406:261–270. https://doi.org/10.1016/s0003-9861(02)00458-7
Nirmaladevi D, Venkataramana M, Srivastava RK et al (2016) Molecular phylogeny, pathogenicity and toxigenicity of Fusarium oxysporum f. sp. lycopersici. Scientific Reports 6:21367. https://doi.org/10.1038/srep21367
Olivain C, Alabouvette C (1999) Process of tomato root colonization by a pathogenic strain of Fusarium oxysporum f. sp. lycopersici in comparison with a non-pathogenic strain. New Phytol 141:497–510. https://doi.org/10.1046/j.1469-8137.1999.00365.x
Pott MB, Effmert U, Piechulla B (2003) Transcriptional and post-translational regulation of S-adenosyl-L-methionine: salicylic acid carboxyl methyltransferase (SAMT) duringStephanotis floribunda flower development. J Plant Physiol 160:635–643. https://doi.org/10.1078/0176-1617-00968
Prihatna C, Barbetti MJ, Barker SJ (2018) A novel tomato Fusarium wilt tolerance gene. Front Microbiol 9:1–11. https://doi.org/10.3389/fmicb.2018.01226
Ranjan A, Ichihashi Y, Sinha NR (2012) The tomato genome: implications for plant breeding, genomics and evolution. Genome Biol 13:167. https://doi.org/10.1186/gb4037
Rep M, Dekker HL, Vossen JH et al (2003a) A tomato xylem sap protein represents a new family of small cysteine-rich proteins with structural similarity to lipid transfer proteins. FEBS Lett 534:82–86. https://doi.org/10.1016/S0014-5793(02)03788-2
Rep M, Dekker HL, Vossen JH et al (2003b) Mass Spectrometric Identification of Isoforms of PR Proteins in Xylem Sap of Fungus-Infected Tomato 1. Plant Physiol 130:904–917. https://doi.org/10.1104/pp.007427.only
Rep M, Van Der Does HC, Meijer M et al (2004) A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol 53:1373–1383. https://doi.org/10.1111/j.1365-2958.2004.04177.x
Rep M, Meijer M, Houterman PM et al (2005) Fusarium oxysporum evades I-3-mediated resistance without altering the matching avirulence gene. Mol Plant Microbe Interact 18:15–23. https://doi.org/10.1094/MPMI-18-0015
Ross JR, Nam KH, D’Auria JC, Pichersky E (1999) S-adenosyl-L-methionine:salicylic acid carboxyl methyltransferase, an enzyme involved in floral scent production and plant defense, represents a new class of plant methyltransferases. Arch Biochem Biophys 367:9–16. https://doi.org/10.1006/abbi.1999.1255
Ruelland E, Campalans A, Selman-Housein G et al (2003) Cellular and subcellular localization of the lignin biosynthetic enzymes caffeic acid-O-methyltransferase, cinnamyl alcohol dehydrogenase and cinnamoyl-coenzyme A reductase in two monocots, sugarcane and maize. Physiol Plant 117:93–99. https://doi.org/10.1034/j.1399-3054.2003.1170112.x
Saikia B, Debbarma J, Maharana J et al (2020) SlHyPRP1 and DEA1, the multiple stress responsive eight-cysteine motif family genes of tomato (Solanum lycopersicum L.) are expressed tissue specifically, localize and interact at cytoplasm and plasma membrane in vivo. Physiol Mol Biol Plants 26:2553–2568. https://doi.org/10.1007/s12298-020-00913-z
Salminen TA, Blomqvist K, Edqvist J (2016) Lipid transfer proteins: classification, nomenclature, structure, and function. Planta 244:971–997. https://doi.org/10.1007/s00425-016-2585-4
Satoh S (2006) Organic substances in xylem sap delivered to above-ground organs by the roots. J Plant Res 119:179–187. https://doi.org/10.1007/s10265-005-0257-8
Subramanian S, Cho U-H, Keyes C, Yu O (2009) Distinct changes in soybean xylem sap proteome in response to pathogenic and symbiotic microbe interactions. BMC Plant Biol 9:119. https://doi.org/10.1186/1471-2229-9-119
Sunitha DCN, N RK, et al (2020) International Journal of Ecology and Environmental Sciences Assessment of tomato (Solanum lycopersicon L.) hybrids for performance and adoptability at Srikakulam District, Andhra Pradesh. Interantional Journal of Ecology and Environmental Sciences 2:317–319
Szklarczyk D, Franceschini A, Wyder S et al (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452. https://doi.org/10.1093/nar/gku1003
Tieman D, Zeigler M, Schmelz E et al (2010) Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J 113–123. https://doi.org/10.1111/j.1365-313X.2010.04128.x
Ueki S, Magori S, Lacroix B, Citovsky V (2013) Transient gene expression in epidermal cells of plant leaves by Biolistic DNA delivery. Methods Mol Biol 940:17–26. https://doi.org/10.1007/978-1-62703-110-3
Upreti R, Thomas P (2015) Root-associated bacterial endophytes from Ralstonia solanacearum resistant and susceptible tomato cultivars and their pathogen antagonistic effects. Front Microbiol 6:1–12. https://doi.org/10.3389/fmicb.2015.00255
van der Does HC, Constantin ME, Houterman PM et al (2019) Fusarium oxysporum colonizes the stem of resistant tomato plants, the extent varying with the R-gene present. Eur J Plant Pathol 154:55–65. https://doi.org/10.1007/s10658-018-1596-3
Walker AM, Sattler SA, Regner M et al (2016) The structure and catalytic mechanism of Sorghum bicolor Caffeoyl-CoA O-methyltransferase. Plant Physiol 172:78–92. https://doi.org/10.1104/pp.16.00845
Wu L, Wang X, Xu R, Li H (2013) Difference between resistant and susceptible maize to systematic colonization as revealed by DsRed-labeled Fusarium verticillioides. The Crop Journal 1:61–69. https://doi.org/10.1016/j.cj.2013.07.004
Yang Q, He Y, Kabahuma M et al (2017) A gene encoding maize caffeoyl-CoA O-methyltransferase confers quantitative resistance to multiple pathogens. Nat Genet 49:1364–1372. https://doi.org/10.1038/ng.3919
Yang J, Wang X, Xie M et al (2019) Proteomic analyses on xylem sap provides insights into the defense response of Gossypium hirsutum against Verticillium dahliae. J Proteomics 213:103599. https://doi.org/10.1016/j.jprot.2019.103599
Yoo S-D, Cho Y-H, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572. https://doi.org/10.1038/nprot.2007.199
Yu C, Cheng C, Su W et al (2014) CELLO2GO: a web server for protein subCELlular LOcalization Prediction with Functional Gene Ontology Annotation. PLoS ONE 9:e99368. https://doi.org/10.1371/journal.pone.0099368
Yue J, Xu W, Ban R et al (2016) PTIR: Predicted Tomato Interactome Resource. Sci Rep 6. https://doi.org/10.1038/srep25047
Zhang G, Zhang W (2019) Protein–protein interaction network analysis of insecticide resistance molecular mechanism in Drosophila melanogaster. Arch Insect Biochem Physiol 100:e21523. https://doi.org/10.1002/arch.21523
Zhang Z, Xin W, Wang S et al (2015) Xylem sap in cotton contains proteins that contribute to environmental stress response and cell wall development. Funct Integr Genomics 15:17–26. https://doi.org/10.1007/s10142-014-0395-y
Zubieta C, Ross JR, Koscheski P et al (2003) Structural basis for substrate recognition in the salicylic acid carboxyl methyltransferase family. Plant Cell 15:1704–1716. https://doi.org/10.1105/tpc.014548.lease
Acknowledgements
The authors acknowledge the Director, CSIR-NEIST Jorhat for the facility and lab space. Authors thank ICAR-IIHR, Bangalore for providing the seeds of tomato cultivars. Authors also acknowledge Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India for PhD program of JD and BS.
Funding
The work was funded by Science and Engineering Research Board (SERB), Govt. of India, Grant Code: SB/S2/RJN-078/2014 as Ramanujan Fellowship and Grant Code: ECR/2016/001288 as Early Career Research Award to C.C.
Author information
Authors and Affiliations
Contributions
CC designed the concept, discussed and corrected the manuscript and involved in overall co-ordination of the project. JD collected the data, performed all the experiments, prepared the figures and tables, analysed the data and drafted the manuscript. BS carried out data analysis, helped in qRT-PCR experiments and corrected the manuscript. DLS, JM, NV, HD and KPA have revised the manuscript, revised the figures, provided critical comments, helped in analysis and corrected the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Debbarma, J., Saikia, B., Singha, D.L. et al. XSP10 and SlSAMT, Fusarium wilt disease responsive genes of tomato (Solanum lycopersicum L.) express tissue specifically and interact with each other at cytoplasm in vivo. Physiol Mol Biol Plants 27, 1559–1575 (2021). https://doi.org/10.1007/s12298-021-01025-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01025-y