Abstract
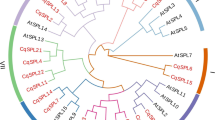
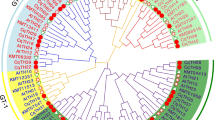
Lim family members play an important role in the regulation of plant cell development and stress response. However, there are few studies on LIM family in quinoa. In this study, we identified nine LIMS (named cqlim01-cqlim09) from quinoa, which were divided into three subfamilies (α Lim1, γ lim2 and δ lim2) according to phylogeny. The differences in gene structure and motif composition among different subfamilies have been observed. In addition, we studied the repetitive events of the members of the family. The Ka/Ks (non synchronous substitution rate / synchronous substitution rate) ratio analysis showed that the repetitive CqLIMs probably experienced purifying selection pressure. Promoter analysis showed that the family genes contained a variety of hormones, stress and tissue-specific expression elements, and protein interactions showed that these genes had actin stabilizing effect. In addition, QRT PCR results showed that all CqLIM genes were positively regulated under three stresses (low temperature, salt and ABA). These results provide a theoretical basis of further study of LIM gene in quinoa.







Similar content being viewed by others
References
Arnaud D, Dejardin A, Leple JC, Lesage-Descauses MC, Pilate G (2007) Genome-wide analysis of LIM gene family in populus trichocarpa, arabidopsis thaliana, and oryza sativa. DNA Res 14:103–116
Arnaud D, Déjardin A, Leplé JC (2012) Expression analysis of LIM gene family in poplar, toward an updated phylogenetic classification. BMC Res Notes 5(1):102
Baltz R, Evrard JL, Domon C, Steinmetz A (1992) A LIM motif is present in a pollen-specific protein. Plant Cell 9:264–268
Baltz R, Schmit AC, Kohnen M, Hentges F, Steinmetz A (1999) Differential localization of the LIM domain protein PLIM-1 in microspores and mature pollen grains from sunflower. Sex Plant Reprod 12(1):60–65
Brière C, Bordel AC, Barthou H, Jauneau A, Steinmetz A, Alibert G, Petitprez M (2003) Is the LIM-domain protein Ha WLIM1 associated with cortical microtubules in sun flower protoplasts. Plant Cell Physiol 44(10):105–1063
Cai XH (2011) Genome-wide analysis of LIM Genes and evolution pattens in Zea mays L. Shandong Agriculture University, Hefei
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4(1):1–21
Chen X, Chen Z, Zhao Y, Cheng BJ, Xiang Y (2014) Genome-wide analysis of soybean HD-Zip gene family and expression profiling under salinity and drought treatments. PLoS ONE 9:e87156
Chen CJ, Chen H, Zhang Y, Thomas HR, Frank MH, He YH (2020) TBtools, a toolkit for biologists integrating various HTS-data handling tools with a user-friendly interface. Mol Plant 13(8):1194–1202
Cheng X, Li G, Muhammad A, Zhang J, Jiang T, Jin Q, Zhao H, Cai Y, Lin Y (2019) Molecular identification, phylogenomic characterization and expression patterns analysis of the LIM (LIN-11, Isl1 and MEC-3 domains) gene family in pear (Pyrus bretschneideri) reveal its potential role in lignin metabolism. Gene 686:237–249
Dawid IB, Breen JJ, Toyama R (1998) LIM domains: multiple roles as adapters and functional modifiers in protein interactions. Trends Genet 14:156–162
Dawid IB, Toyama R, Taira M (1995) Lim domain proteins. Comptes Rendus de l’Académie des Sciences de Paris 318:295–306
Day B, Henty JL, Porter KJ, Staiger CJ (2011) The pathogen-actin connection: a platform for defense signaling in plants. Annu. Rev. Phytopathol 49(1):483–506
Eliasson A, Gass N, Mundel C, Baltz R, Krauter R, Evrard JL, Steinmetz A (2000) Molecular and expression analysis of a LIM protein gene family from flowering plants. Mol Gen Genet 264:257–267
Fan B, Liu MX, Li HJ, Zhang X, Wang XM, Cui XY (2015) Bioinformatics analysis of AAP gene family in Arabidopsis thaliaha. Chem Life 36(3):372–378
Gabaldón T, Koonin EV (2013) Functional and evolutionary implications of gene orthology. Nat Rev Genet 14:360–366
Gao X, Chen Y, Chen M, Wang S, Wen X, Zhang S (2018) Identification of key candidate genes and biological pathways in bladder cancer. PeerJ 6(4):e6036
Gardy JL, Laird MR, Chen F, Rey S, Walsh CJ, Ester M (2005) PSORTb v.2.0: expanded prediction of bacterial protein subcellular localization and insights gained from comparative proteome analysis. Bioinformatics 21(5):617–623
Graf BL, Rojas-Silva P, Rojo LE, Delatorre-Herrera J, Baldeón ME, Raskin I (2015) Innovations in health value and functional food development of Quinoa (Chenopodium quinoa Willd.). Compr Rev Food Sci Food Saf 14(4):431–445
Han L-B, Li Y-B, Wang H-Y, Wu X-M, Li C-L, Luo M, Wu S-J, Kong Z-S, Pei Y, Jiao G-L, Xia G-X (2013) The dual functions of WLIM1a in cell elongation and secondary wall formation in developing cotton fibers. Plant Cell 25:4421–4438
Hen X, Song R, Yu M, Sui J, Wang J, Qiao L (2015) Cloning and functional analysis of the chitinase gene promoter in peanut. Genet Mol Res 14(4):12710
Hu B, Jin J, Guo AY, Zhang H, Luo J, Gao G (2015) GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31(8):1296–1297
Jong-In P, Nasar UA, Hee-Jeong J (2014) Identification and characterization of LIM gene family in Brassica rapa. BMC Genom 15(1):641
Kawaoka A, Ebinuma H (2001) Transcriptional control of lignin biosynthesis by tobacco LIM protein. Phytochemistry 57(7):1149–1157
Kawaoka A, Kaothien P, Yoshida K, Endo S, Yamada K, Ebinuma H (2000) Functional analysis of tobacco LIM protein NtLIM1 involved in lignin biosynthesis. Plant J 22:289–301
Kaufmann K, Melzer R, Theiβen G (2005) MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347(2):183–198
Kumar S, Stecher G, Tamura K (2006) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mole Biol Evol 33(7):1870–1874
Khatun K, Robin AK, Park JI, Ahmed NU, Kim CK, Lim KB, Kim MB, Lee DJ, Nou IS, Chung MY (2016) Genome-wide identification, characterization and expression profiling of LIM family genes in Solanum lycopersicum L. Plant Physiol Biochem 108:177–190
Kim JS, Mizoi J, Yoshida T, Fujita Y, Nakajima J, Ohori T, Todaka D, Nakashima K, Hirayama T, Shinozaki K, Yamaguchi-Shinozaki K (2011) An ABRE promoter sequence is involved in osmotic stress-responsive expression of the DREB2A gene, which encodes a transcription factor regulating drought-inducible genes in Arabidopsis. Plant Cell Physiol 52:2136–2146
Labalette C, Nouet Y, Levillayer F, Colnot S, Chen J, Claude V, Huerre M, Perret C, Buendia MA, Wei Y (2010) Deficiency of the LIM-only protein FHL2 reduces intestinal tumorigenesis in Apc mutant mice. PLoS ONE 5:e10371
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van PY (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucl Acids Res 30:325–327
Li RX, Zhu FD, Duan D (2020) Function analysis and stress-mediated cis-element identification in the promoter region of VqMYB15. Plant Signal Behav 15(7):1773664
Li Y, Jiang J, Li L, Wang XL, Wang NN, Li DD, Li XB (2013) A cotton LIM domain-containing protein (ghwlim5) is involved in bundling actin filaments. Plant Physiol Biochem 66:34–40
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR. Method 25:402–408
Lynch M, Conery JS (2003) The evolutionary demography of duplicate genes. J Struct Funct Genom 3:35–44
Maul RS, Song Y, Amann KJ, Gerbin SC, Pollard TD, Chang DD (2003) EPLIN regulates actin dynamics by cross-linking and stabilizing filaments. J Cell Biol 160(6):399–407
Moes D, Gatti S, Hoffmann C, Dieterle M, Moreau F, Neumann K, Schumacher M, Diederich M, Grill E, Shen WH (2012) A LIM domain protein from tobacco involved in actin-bundling and histone gene transcription. Mole Plant 6(2):483–502
Mundel C, Baltz R, Eliasson A, Bronner R, Grass N, Kruter R, Evrard JL, Steinmetz A (2000) A LIM-domain protein from sunflower is localized to the cytoplasm and/or nucleus in a wide variety of tissues and is associated with the phragmoplast in dividing cells. Plant Mol Biol 42(2):291–302
Pant P, Iqbal Z, Pandey BK, Sawant SV (2018) Genome-wide comparative and evolutionary analysis of calmodulin-binding transcription activator (CAMTA) family in Gossypium species. Sci Rep 8(1):5573
Papuga J, Hoffmann C, Dieterle M, Moes D, Moreau F, Tholl S, Steinmetz A, Thomas C (2010) Arabidopsis LIM proteins: a family of actin bundlers with distinct expression patterns and modes of regulation. Plant Cell 22:3034–3052
Papuga J, Thomas C, Dieterle M, Moreau F, Steinmetz A (2009) Arabidopsis LIM domain proteins involved in actin bundling exhibit different modes of regulation. FEBS J 276:245
Park JI, Ahmed NU, Jung HJ, Arasan SK, Chung MY, Cho YG, Watanabe M, Nou IS (2014) Identification and characterization of LIM gene family in brassica rapa. BMC Genom 15:641
Sala S, Ampe C (2018) An emerging link between lim domain proteins and nuclear receptors. Cell Mol Life Sci 75:1959–1971
Shaul O (2017) How introns enhance gene expression. Int J Biochem Cell Biol 91:145–155
Shen XX, Salichos L, Rokas A (2016) A genome-scale investigation of how sequence, function, and tree-based gene properties influence phylogenetic inference. Genome Biol Evol 8:2565–2580
Srivastava V, Verma PK (2017) The plant LIM proteins: unlocking the hidden attractions. Planta 246:365–375
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucl Acids Res 34:W609–W612
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J (2015) STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucl Acids Res 43:D447–D452
Steven DC (1996) Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J 10(1):1–8
Tian F, Yang DC, Meng YQ, Jin JP, Gao G (2019) PlantRegMap: charting functional regulatory maps in plants. Nucl Acids Res. https://doi.org/10.1093/nar/gkz1020
Thomas C, Dieterle M, Gatti S, Hoffmann C, Moreau F, Papuga J, Steinmetz A (2008) Actin bundling via LIM domains. Plant Signal Behav 3(5):3200–3211
Thomas C, Hoffmann C, Dieterle M, Van Troys M, Ampe C, Steinmetza A (2006) Tobacco wlim1 is a novel f-actin binding protein involved in actin cytoskeleton remodeling. Plant Cell 18:2194–2206
Tohge T, Fernie AR (2010) Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc 5:1210–1227
Tran TC, Singleton C, Fraley TS, Greenwood JA (2005) Cysteine-rich protein 1(CRP1) regulates actin filament bundling. BMC Cell Biol 6:45
Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290:2114–2117
Wang C, Zhang L, Huang R (2011a) Cytoskeleton and plant salt stress tolerance. Plant Signal Behav 6(1):29–31
Wang Y, Liu GJ, Yan XF, Wei ZG, Xu ZR (2011b) MeJA-inducible expression of the heterologous jaz2 promoter from arabidopsis in populus trichocarpa protoplasts. Plant Dis 118:69–74
Wang Y, Wang C, Rajaofera M, Jayne N, Zhu L, Liu WB, Zheng FC, Miao WG (2020) WY7 is a newly identified promoter from the rubber powdery mildew pathogen that regulates exogenous gene expression in both monocots and dicots. PLoS ONE 15(6):e0233911
Wang Y, Wang X, Paterson AH (2012) Genome and gene duplications and gene expression divergence: a view from plants. Ann N Y Acad Sci 1256(1):1–14
Wasteneys GO, Yang Z (2004) The cytoskeleton becomes multidisciplinary. Plant Physiol 136(4):3853–3854
Way JC, Chalfie M (1988) mec-3, a homeobox-containing gene that specifies differentiation of the touch receptor neurons in C. elegans. Cell 54:5–16
Yang S, Zhu G, Wang C, Chen L, Song Y, Wang J (2017) Regulation of secondary xylem formation in young hybrid poplars by modifying the expression levels of the PtaGLIMa gene. Mol Breed 37:37–40
Yang R, Chen M, Sun JC (2019) Genome-Wide Analysis of LIM Family Genes in Foxtail Millet (Setaria italica L.) and Characterization of the Role of SiWLIM2b in Drought Tolerance. Int J Mol Sci 20(6):1303–1309
Ye J, Xu M (2012) Actin bundler PLIM2s are involved in the regulation of pollen development and tube growth in Arabidopsis. J Plant Physiol 169(5):516–522
Zhang X, Cheng T, Wang G, Yan Y (2013) Cloning and evolutionary analysis of tobacco MAPK gene family. Mol Biol Rep 40:1407–1415
Zhang Z, Li J, Zhao XQ, Wang J, Wong GKS, Yu J (2006) KaKs_calculator: calculating Ka and Ks through model selection and model averaging. Genom Proteom Bioinform 4:259–263
Zhang H, Gao S, Lercher MJ, Hu S, Chen WH (2012) EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucl Acids Res 4:W569–W572
Zhao M, He L, Gu Y, Wang Y, Chen Q, He C (2014) Genome-wide analyses of a plant-specific LIM-domain gene family implicate its evolutionary role in plant diversification. Genome Biol Evol 6:1000–1012
Zheng Q, Zhao Y (2007) The diverse biofunctions of LIM domain proteins: determined by subcellular localization and protein-protein interaction. Biol Cell 99:489–502
Acknowledgements
The authors thank all contributors to this research for their help. The research was financially supported by National Natural Science Foundation of China, Gansu Provincial Key Laboratory of Aridland Crop Science.
Funding
The research was financially supported by National Natural Science Foundation of China (no. 32060401).
Author information
Authors and Affiliations
Contributions
Data curation, X.W; Funding acquisition, X.W. and C.Z.; Methodology, C.Z., X.W., and X.Z.; Project administration,X.W.; Resources, L.H.; Supervision,X.Z and X.W.; Writing—original draft, B.W.; Writing—review and editing, X.Z and X.W.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, X., Wang, B., Wang, X. et al. Genome-wide identification, characterization and expression analysis of the LIM transcription factor family in quinoa. Physiol Mol Biol Plants 27, 787–800 (2021). https://doi.org/10.1007/s12298-021-00988-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-00988-2