Abstract
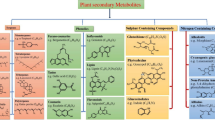
Late leaf spot (LLS) caused by fungi Passalora personata is generally more destructive and difficult to control than early leaf spot. The aim of this study was to decipher biochemical defense mechanism in groundnut genotypes against P. personata by identifying resistance specific biomarkers and metabolic pathways induced during host–pathogen interaction. Metabolomics of non-infected and infected leaves of moderately resistant (GPBD4 and ICGV86590), resistant (KDG128 and RHRG06083) and susceptible (GG20, JL24 and TMV2) genotypes was carried out at 5 days after infection (65 days after sowing). Non-targeted metabolite analysis using GC–MS revealed total 77 metabolites including carbohydrates, sugar alcohols, amino acids, fatty acids, polyamines, phenolics, terpenes and sterols. Variable importance in projection (VIP) measure of partial least squares-discriminant analysis (PLS-DA) showed that resistant and moderately resistant genotypes possessed higher intensities of ribonic acid, cinnamic acid, malic acid, squalene, xylulose, galactose, fructose, glucose, β-amyrin and hydroquinone while susceptible genotypes had higher amount of gluconic acid 2-methoxime, ribo-hexose-3-ulose and gluconic acid. Heat map analysis showed that resistant genotypes had higher intensities of β-amyrin, hydroquinone in non-infected and malic acid, squalene, putrescine and 2,3,4-trihydroxybutyric acid in infected leaves. Dendrogram analysis further separated resistant genotypes in the same cluster along with infected moderately resistant genotypes. The most significant pathways identified are: linoleic acid metabolism, flavone and flavonol biosynthesis, cutin, suberin and wax biosynthesis, pentose and glucuronate interconversions, starch and sucrose metabolism, stilbenoid biosynthesis and ascorbate and aldarate metabolism. Targeted metabolite analysis further confirmed that resistant genotypes possessed higher content of primary metabolites sucrose, glucose, fructose, malic acid and citric acid. Moreover, resistant genotypes possessed higher content of salicylic, coumaric, ferulic, cinnamic, gallic acid (phenolic acids) and kaempferol, quercetin and catechin (flavonols). Thus metabolites having higher accumulation in resistant genotypes can be used as biomarkers for screening of LSS resistant germplasm. These results unravel that higher amount of primary metabolites leads to stimulate the accumulation of more amounts of secondary metabolites such as phenolic acid, flavanols, stilbenes and terpenoids (squalene and β-amyrin) biosynthesis which are ultimately involved in defense mechanism against LLS pathogen.







Similar content being viewed by others
References
Alkan N, Friedlander G, Ment D, Prusky D, Fluhr R (2015) Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol 205:801–881
Augustyn WA, Regnier T, Combrinck S, Botha BM (2014) Metabolic profiling of mango cultivars to identify biomarkers for resistance against Fusarium infection. Phytochem Lett 10:civ–cx
Balmer A, Pastor V, Glauser G, Mauch-Mani B (2018) Tricarboxylates induce defense priming against bacteria in Arabidopsis thaliana. Front Plant Sci 9:1221
Bao G, Zhuo C, Qian C, Xiao T, Guo Z, Lu S (2016) Co-expression of NCED and ALO improves vitamin C level and tolerance to drought and chilling in transgenic tobacco and stylo plants. Plant Biotechnol J 14:206–214
Blée E (2003) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7:315–322
Bollina V, Kushalappa AC, Choo TM, Dion Y, Rioux S (2011) Identification of metabolites related to mechanisms of resistance in barley against Fusarium graminearum, based on mass spectrometry. Plant Mol Biol 77:355–370
Bolton MD (2009) Primary metabolism and plant defense—fuel for the fire. Mol Plant-Microbe Interact 22:487–497
Buschhaus C, Jetter R (2011) Composition differences between epicuticular and intracuticular wax substructures: how do plants seal their epidermal surfaces? J Exp Biol 62:841–853
Chaturvedi R, Shah J (2007) Salicylic acid in plant disease resistance. In: Hayat S, Ahmad A (eds) Salicylic acid: a plant hormone. Springer, Dordrecht, pp 335–370
Chitarrini G, Soini E, Riccadonna S, Franceschi P, Zulini L, Masuero D, Vecchione A, Stefanini M, Di Gaspero G, Mattivi F, Vrhovsek U (2017) Identification of biomarkers for defense response to Plasmapara viticola in a resistant grape variety. Front Plant Sci 8:1524
Chong IG, Jun CH (2005) Performance of some variable selection methods when multicollinearity is present. Chemometr Intell Lab 78:103–112
Chong J, Wishart DS, Xia J (2019) Using MetaboAnalyst 4.0 for comprehensive and integrative metabolomics data analysis. Curr Protoc Bioinform 68:e86
Daniele E, Dommes J, Hausman JF (2003) Carbohydrates and resistance to Phytophthora infestans in potato plants. Acta Physiol Plant 25:171–178
Del Río JA, Báidez AG, Botía JM, Ortuño A (2003) Enhancement of phenolic compounds in olive plants (Olea europaea L.) and their influence on resistance against Phytophthora sp. Food Chem 83:75–78
Delaney TP, Uknes S, Vernooij B, Friedrich L, Weymann K, Negrotto D, Gaffney T (1994) A central role of salicylic acid in plant disease resistance. Science 266:1247–1250
De Oliveira CS, Lião LM, Alcantara GB (2019) Metabolic response of soybean plants to Sclerotinia sclerotiorum infection. Phytochemistry 167:112099
Engelsdorf T, Horst RJ, Pröls R, Pröschel M, Dietz F, Hückelhoven R, Voll LM (2013) Reduced carbohydrate availability enhances the susceptibility of arabidopsis toward: Colletotrichum higginsianum. Plant Physiol 162:225–238
Fathima A, Rao JR (2016) Selective toxicity of Catechin—a natural flavonoid towards bacteria. Appl Microbiol Biotechnol 100:6395–6402
Fernie AR, Schauer N (2009) Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet 25:39–48
Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3:408–414
Grichar WJ, Besler BA, Jaks AJ (1998) Groundnut (Arachis hypogaea L.) cultivar response to leaf spot disease development under four disease management programs. Peanut Sci 25:35–39
Gromski PS, Muhamadali H, Ellis DI, Xu Y, Correa E, Turner ML, Goodacre RA (2015) Tutorial review: metabolomics and partial least squares-discriminant analysis–A marriage of convenience or a shotgun wedding. Anal Chim Acta 879:10–23
Hamzehzarghani H, Vikram A, Abu-Nada Y, Kushalappa AC (2016) Tuber metabolic profiling of resistant and susceptible potato varieties challenged with Phytophthora infestans. Eur J Plant Pathol 145:277–287
Hamzehzarghani H, Paranidhara V, Abu-Nada Y, Kushalappa AC, Dion Y, Rioux S, Comeau A, Yaylayan V, Marshall W (2008) Metabolic profiling coupled with statistical analyses for potential high throughput screening of quantitative resistance to fusarium head blight in wheat cultivars. Can J Plant Pathol 30:24–36
Islam MT, Lee BR, Das PR, La VH, Leea H, Jungc WJ, Baed DW, Kim TH (2019) p-Coumaric acid induces jasmonic acid-mediated phenolic accumulation and resistance to black rot disease in Brassica napus. Physiol Mol Plant P 106:270–275
Jadhav PR, Mahatma MK, Mahatma L, Jha S, Parekh VB, Khandelwal V (2013) Expression analysis of key genes of phenylpropanoid pathway and phenol profiling during Ricinus communis–Fusarium oxysporum f. sp. ricini interaction. Ind Crop Prod 50:456–461
Jain R, Jha S, Adhikary H, Kumar P, Parekh V, Jha A, Mahatma MK, Kumar GN (2014) Isolation and molecular characterization of arsenite-tolerant Alishewanella sp. GIDC-5 originated from industrial effluents. Geomicrobiol J 31:82–90
Kachroo A, Kachroo P (2009) Fatty acid–derived signals in plant defense. Ann Rev Phytopathol 47:153–176
Kemen AC, Honkanen S, Melton RE, Findlay KC, Mugford ST, Hayashi K, Haralampidis K, Rosser SJ, Osbourn A (2014) Investigation of triterpene synthesis and regulation in oats reveals a role for β-amyrin in determining root epidermal cell patterning. Proc Natl Acad Sci USA 111:8679–8684
Khatediya NK, Parmar DV, Mahatma MK, Pareek M (2018) Increased accumulation of phenolic metabolites in groundnut (Arachis hypogaea L.) genotypes contribute to defense against Sclerotium rolfsii infection. Arch Phytopath Plant Protec 51:530–549
Kouzai Y, Kimura M, Watanabe M, Kusunoki K, Osaka D, Suzuki T, Matsui H, Yamamoto M, Ichinose Y, Toyoda K, Matsuura T, Mori IC, Hirayama T, Minami E, Nishizawa Y, Inoue K, Onda Y, Mochida K, Noutoshi Y (2018) Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytol 217:771–783
Kumar R, Bohra A, Pandey AK, Pandey MK, Kumar A (2017) Metabolomics for plant improvement: status and prospects. Front Plant Sci 8:1302
Lecompte F, Nicot PC, Ripoll J, Abro MA, Raimbault AK, Lopez-Lauri F, Bertin N (2017) Reduced susceptibility of tomato stem to the necrotrophic fungus Botrytis cinerea is associated with a specific adjustment of fructose content in the host sugar pool. Ann Bot 119:931–943
Li W, Li C, Sun J, Peng M (2017) Metabolomic, biochemical, and gene expression analyses reveal 628 the underlying responses of resistant and susceptible banana species during early infection 629 with Fusarium oxysporum f. sp. cubense. Plant Dis 101:534–543
Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1:1–10
Mahatma MK, Thawait LK, Jadon KS, Rathod KJ, Sodha KH, Bishi SK, Thirumalaisamy PP, Golakiya BA (2019) Distinguish metabolic profiles and defense enzymes in Alternaria leaf blight resistant and susceptible genotypes of groundnut. Physiol Mol Biol Plants 25:1395–1405
Mahatma MK, Thawait LK, Jadon KS, Thirumalaisamy PP, Bishi SK, Jadav JK, Khatediya N, Golakiya BA (2018) Metabolic profiles of groundnut (Arachis hypogaea L.) genotypes differing in Sclerotium rolfsii reaction. Eur J Plant Pathol 151:463–474
Mahatma MK, Thawait LK, Bishi SK, Khatediya N, Rathnakumar AL, Lalwani HB, Misra JB (2016) Nutritional composition and antioxidant activity of Spanish and Virginia groundnuts (Arachis hypogaea L.): a comparative study. J Food Sci Technol 53:2279–2286
Morkunas I, Formela M, Marczak L, Stobiecki M, Bednarski W (2013) Themobilization of defence mechanisms in the early stages of pea seed germination against Ascochyta pisi. Protoplasma 250:63–75
Osbourn AE (1996) Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 8:1821–1831
Padmavati M, Sakyhivel N, Thara KV, Reddy AR (1997) Differential sensitivity of rice pathogens to growth inhibition by flavonoids. Phytochemistry 46:499–502
Pandey D, Rajendran SRCK, Gaur M, Sajeesh PK, Kumar A (2016) Plant defense signaling and responses against necrotrophic fungal pathogens. J Plant Growth Regul 35:1159–1174
Papadopoulou K, Melton RE, Leggett M, Daniels MJ, Osbourn AE (1999) Compromised disease resistance in saponin-deficient plants. Proc Natl Acad Sci USA 96:12923–12928
Raval SS, Mahatma MK, Chakraborty K, Bishi SK, Singh AL, Rathod KJ, Jadav JK, Sanghani JM, Mandavia MK, Gajera HP, Golakiya BA (2018) Metabolomics of groundnut (Arachis hypogaea L.) genotypes under varying temperature regimes. Plant Growth Regul 84:493–505
Skłodowska M, Mikiciński A, Wielanek M, Kuźniak E, Sobiczewski P (2018) Phenolic profiles in apple leaves and the efficacy of selected phenols against fire blight (Erwinia amylovora). Eur J Plant Pathol 151:213–228
Steinfath M, Strehmel N, Peters R, Schauer N, Groth D, Hummel J, Steup M, Selbig J, Kopka J, Geigenberger P, van Dongen JT (2010) Discovering plant metabolic biomarkers for phenotype prediction using an untargeted approach. Plant Biotechnol J 8:900–911
Subrahmanyam P, McDonald D, Waliyar F, JReddy L, Nigam SN, Gibbons RW, Ramanatha Rao V, Singh AK, Pande S, Reddy PM, Subba Rao PV (1995) Screening methods and sources of resistance to rust and late leaf spot of groundnut. Information Bulletin no. 47. International Crops Research Institute for the Semi-Arid Tropics Patanchem 502 324, Andhra Pradesh, India.
Swami RM, Mahatma MK, Parekh MJ, Kalariya KA, Mahatma L (2015) Alteration of metabolites and polyphenol oxidase activity in wilt resistant and susceptible pigeon pea genotypes during Fusarium udum infection. Indian J Agric Biochem 28:18–23
Tugizimana F, Steenkamp P, Piater L, Dubery I (2016) A conversation on data mining strategies in LC-MS untargeted metabolomics: pre-processing and pre-treatment steps. Metabolites 6:1–18
Ullah C, Unsicker SB, Fellenberg C, Constabel CP, Schmidt A, Gershenzon J, Hammerbachera A (2017) Flavan-3-ols are an effective chemical defense against rust infection. Plant Physiol 175:1560–1578
Vadivel AK (2015) Gel-based proteomics in plants: time to move on from the tradition. Front Plant Sci 6:369
Vogt T (2010) Phenylpropanoid biosynthesis. Mol Plant 3:2–20
Wang X, Kong L, Zhi P, Chang C (2020) Update on cuticularwax biosynthesis and its roles in plant disease resistance. Int J Mol Sci 21:5514
Wen W, Li K, Alseekh S, Omranian N, Zhao L, Zhou Y, Xiao Y, Jin M, Yang N, Liu H, Florian A, Li W, Pan Q, Nikoloski Z, Yan J, Fernie AR (2015) Genetic determinants of the network of primary metabolism and their relationships to plant performance in a maize recombinant inbred line population. Plant Cell 27:1839–1856
Xia J, Wishart D (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6:743–760
Yogendra KN, Kushalappa AC, Sarmiento F, Rodriguez E, Mosquera T (2015) Metabolomics deciphers quantitative resistance mechanisms in diploid potato clones against late blight. Funct Plant Biol 42:284–298
Yogendra KN, Pushpa D, Mosa KA, Kushalappa AC, Murphy A, Mosquera T (2014) Quantitative resistance in potato leaves to late blight associated with induced hydroxycinnamic acid amides. Funct Integr Genomic 14:285–298
Zhang YJ, Fernie AR (2018) On the role of the tricarboxylic acid cycle in plant productivity. J Integr Plant Biol 60:1199–1216
Zhang Z, Qin G, Li B, Tian S (2015) Effect of cinnamic acid for controlling gray mold on table grape and its possible mechanisms of action. Curr Microbiol 71:396–402
Ziv C, Zhao Z, Gao YG, Xia Y (2018) Multifunctional roles of plant cuticle during plant-pathogen interactions. Front Plant Sci 9:1088
Zivy M, Wienkoop S, Renaut J, Pinheiro C, Goulas E, Carpentier S (2015) The quest for tolerant varieties: the importance of integrating “omics” techniques to phenotyping. Front Plant Sci 6:448
Acknowledgements
Authors are thankful to the Director, ICAR-Directorate of Groundnut Research, Junagadh, India for providing all the necessary facilities and support to completing this experiment.
Funding
This work was carried out under Institute Project. External funning was not received for this work.
Author information
Authors and Affiliations
Contributions
MKM conceived the study, performed and designed the experiments and drafted the manuscript. LKT carried out sugar and organic acid measurements and phenolics and metabolite extraction. KSJ, TPP, and NK carried out the pot and field experiments to screen groundnut genotypes KSJ and TPP also performed inoculation of disease. KJR carried out untargeted metabolites analysis by GC–MS and phenolics profiling by LC–MS/MS and data processing, SKB and AV performed the statistical analysis and interpretation of results of targeted metabolites. BAG contributed to interpretation of the data and editing of the MS.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mahatma, M.K., Thawait, L.K., Jadon, K.S. et al. Metabolic profiling for dissection of late leaf spot disease resistance mechanism in groundnut. Physiol Mol Biol Plants 27, 1027–1041 (2021). https://doi.org/10.1007/s12298-021-00985-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-00985-5