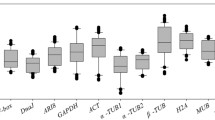
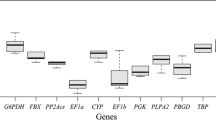
Abstract
Haloxylon ammodendron plays an important role in maintaining the structure and function of the entire ecosystem where it grows. No suitable reference genes have been reported in H. ammodendron plants to date. In this study, a total of 8 reference genes (18S, ACT1, ACT7, UBC18, TUA5, GAPDH, EF-1α and UBQ10) were selected from the available trancriptome database, and the expression stability of these 8 candidate genes was validated under different abiotic stress with three different statistical algorithms (geNorm, NormFinder and BestKeeper). The results produced from different models were in agreement with each other essentially: 18S and TUA5 were the most stable genes under drought stress, 18S, the most stable gene under heat stress and mechanical damage, ACT7 and UBC18, stable under salt stress while TUA5 and GAPDH expressed constantly under mechanical damage, and ACT1 expressed steadily under cold conditions. Expression profiles of several stress response genes, including FT-5, FT-9, DREB2A and DREB2C, were further confirmed with various candidate reference genes. None of the candidate genes showed a constant expression among all tested samples. Hence, it’s essential to use more than one reference gene in order to guarantee the accuracy of quantitative real-time PCR. The results of this study will contribute to the accuracy and reliability in transcripts quantification, which is of significance to transcription-based studies and applications in this important shrub H. ammodendron.




Similar content being viewed by others
References
Amil-Ruiz F, Garrido-Gala J, Blanco-Portales R, Folta KM, Muñoz-Blanco J, Caballero JL (2013) Identification and validation of reference genes for transcript normalization in strawberry (Fragaria × ananassa) defense responses. PLoS ONE 8:e70603. https://doi.org/10.1371/journal.pone.0070603
Andersen CL, Jensen JL, Ørntoft TF (2004) Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Can Res 64:5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496
Bao W, Qu Y, Shan X, Wan Y (2016) Screening and validation of housekeeping genes of the root and cotyledon of Cunninghamia lanceolata under abiotic stresses by using quantitative real-time PCR. Int J Mol Sci 17:1198. https://doi.org/10.3390/ijms17081198
Bustin SA (2002) Quantification of mRNA using real-time reverse transcription PCR (RTPCR): trends and problems. J Mol Endocrinol 29:23–39. https://doi.org/10.1677/jme.0.0290023
Chen IC, Hill JK, Ohlemüller R, Roy DB, Thomas CD (2011) Rapid range shifts of species associated with high levels of climate warming. Science 333:1024–1026. https://doi.org/10.1126/science.1206432
Crismani W, Baumann U, Sutton T, Shirley N, Webster T, Spangenberg G, Langridge P, Able JA (2006) Microarray expression analysis of meiosis and microsporogenesis in hexaploid bread wheat. BMC Genomics 7:267. https://doi.org/10.1186/1471-2164-7-267
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17. https://doi.org/10.1104/pp.105.063743
Derveaux S, Vandesompele J, Hellemens J (2010) How to do successful gene expression analysis using real-time PCR. Methods 50:227–230. https://doi.org/10.1016/j.ymeth.2009.11.001
Ding Y, Liu N, Virlouvet L, Riethoven JJ, Fromm M, Avramova Z (2013) Four distinct types of dehydration stress memory genes in Arabidopsis thaliana. BMC Plant Biol 13:229. https://doi.org/10.1186/1471-2229-13-229
Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345:646–651. https://doi.org/10.1016/j.bbrc.2006.04.140
Le DT, Aldrich DL, Valliyodan B, Watanabe Y, Ha CV, Nishiyama R, Guttikonda SK, Quach TN, Gutierrez-Gonzalez JJ, Tran LS, Nguyen HT (2012) Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE 7:e46487. https://doi.org/10.1371/journal.pone.0046487
Li W, Zhang L, Zhang Y, Wang G, Song D, Zhang Y (2017) Selection and validation of appropriate reference genes for quantitative real-time PCR normalization in staminate and perfect flowers of andromonoecious Taihangia rupestris. Front Plant Sci 8:729. https://doi.org/10.3389/fpls.2017.00729
Lin YL, Lai ZX (2010) Reference genes selection for qPCR analysis during somatic embryo genesis in longan tree. Plant Sci 178:359–365. https://doi.org/10.1016/j.plantsci.2010.02.005
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔC(T)) method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Long Y, Zhang J, Tian X, Wu S, Zhang Q, Zhang J, Dang Z, Pei XW (2014) De novo assembly of the desert tree Haloxylon ammodendron (C. A. Mey.) based on RNA-Seq data provides insight into drought response, gene discovery and marker identification. BMC Genomics 15:1111. https://doi.org/10.1186/1471-2164-15-1111
Ma R, Xu S, Zhao Y, Xia B, Wang R (2016) Selection and validation of appropriate reference genes for quantitative Real-Time PCR Analysis of gene expression in Lycoris aurea. Front Plant Sci 7:536. https://doi.org/10.3389/fpls.2016.00536
Manoharan RK, Han JS, Vijayakumar H, Subramani B, Thamilarasan SK, Park JI, Nou IS (2016) Molecular and functional characterization of FLOWERING LOCUS T homologs in Allium cepa. Molecules 21:217. https://doi.org/10.3390/molecules21020217
Maroufi A, Van Bockstaele E, De Loose M (2010) Validation of reference genes for gene expression analysis in chicory (Cichorium intybus) using quantitative real-time PCR. BMC Mol Biol 11:15. https://doi.org/10.1186/1471-2199-11-15
Oono Y, Yazawa T, Kawahara Y, Kanamori H, Kobayashi F, Sasaki H, Mori S, Wu J, Handa H, Itoh T, Matsumoto T (2014) Genome-wide transcriptome analysis reveals that cadmium stress signaling controls the expression of genes in drought stress signal pathways in rice. PLoS ONE 9:e96946. https://doi.org/10.1371/journal.pone.0096946
Perez LJ, Rios L, Trivedi P, D’Souza K, Cowie A, Nzirorera C, Webster D, Brunt K, Legare JF, Hassan A, Kienesberger PC, Pulinilkunnil T (2017) Validation of optimal reference genes for quantitative real time PCR in muscle and adipose tissue for obesity and diabetes research. Sci Rep 7:3612. https://doi.org/10.1038/s41598-017-03730-9
Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Bestkeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. https://doi.org/10.1023/B:BILE.0000019559.84305.47
Piron Prunier F, Chouteau M, Whibley A, Joron M, Llaurens V (2016) Selection of valid reference genes for reverse transcription quantitative PCR analysis in Heliconius numata (Lepidoptera: Nymphalidae). J Insect Sci 16:50. https://doi.org/10.1093/jisesa/iew034
Pyankov VI, Black CC Jr, Artyusheva EG, Voznesenskaya EV, Ku MSB, Edwards GE (1999) Features of photosynthesis in Haloxylon species of Chenopo-diaceae that are dominant plants in Central Asia desert. Plant Cell Physiol 40:125–134. https://doi.org/10.1093/oxfordjournals.pcp.a029519
Radonić A, Thulke S, Mackay IM, Landt O, Siegert W, Nitsche A (2004) Guideline to reference gene selection for quantitative real-time PCR. Biochem Biophys Res Commun 313:856–862. https://doi.org/10.1016/j.bbrc.2003.11.177
Reddy PS, Reddy DS, Sharma KK, Bhatnagar-Mathur P, Vadez V (2015) Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene 1:35–42. https://doi.org/10.1016/j.plgene.2015.02.001
Ryu JY, Park CM, Seo PJ (2011) The floral repressor BROTHER OF FT AND TFL1 (BFT) modulates flowering initiation under high salinity in Arabidopsis. Mol Cells 32:295–303. https://doi.org/10.1007/s10059-011-0112-9
Shavrukov Y, Baho M, Lopato S, Langridge P (2016) The TaDREB3 transgene transferred by conventional crossings to different genetic backgrounds of bread wheat improves drought tolerance. Plant Biotechnol J 14:313–322. https://doi.org/10.1111/pbi.12385
Sheng Y, Zheng W, Pei K, Ma K (2005) Genetic variation within and among populations of a dominant desert tree Haloxylon ammodendron (Amaranthaceae) in China. Ann Bot 96:245–252. https://doi.org/10.1093/aob/mci171
Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 3:RESEARCH0034. https://doi.org/10.1186/gb-2002-3-7-research0034
Volkov RA, Panchuk II, Schöffl F (2003) Heat-stress-dependency and developmental modulation of gene expression: the potential of house-keeping genes as internal standards in mRNA expression profiling using real-time RT-PCR. J Exp Bot 54:2343–2349. https://doi.org/10.1093/jxb/erg244
Wan H, Zhao Z, Qian C, Sui Y, Malik AA, Chen J (2010) Selection of appropriate reference genes for gene expression studies by quantitative real-time polymerase chain reaction in cucumber. Anal Biochem 399:257–261. https://doi.org/10.1016/j.ab.2009.12.008
Wei L, Miao H, Zhao R, Han X, Zhang T, Zhang H (2013) Identification and testing of reference genes for Sesame gene expression analysis by quantitative real-time PCR. Planta 237:873–889. https://doi.org/10.1007/s00425-012-1805-9
Wu H, Lv H, Li L, Liu J, Mu S, Li X, Gao J (2015) Genome-wide analysis of the AP2/ERF transcription factors family and the expression patterns of DREB genes in Moso Bamboo (Phyllostachys edulis). PLoS ONE 10:e0126657. https://doi.org/10.1371/journal.pone.0126657
Wu J, Zhang H, Liu L, Li W, Wei Y, Shi S (2016) Validation of reference genes for RT-qPCR studies of gene expression in preharvest and postharvest Longan Fruits under different experimental conditions. Front Plant Sci 7:780. https://doi.org/10.3389/fpls.2016.00780
Yu T, Ren C, Zhang J, He X, Ma L, Chen Q, Qu Y, Shi S, Zhang H, Ma H (2012) Effect of high desert surface layer temperature stress on Haloxylon ammodendron (C.A. Mey.) Bunge. Flora 207:572–580. https://doi.org/10.1016/j.flora.2012.06.008
Yuan M, Lu Y, Zhu X, Wan H, Shakeel M, Zhan S, Jin BR, Li J (2014) Selection and evaluation of potential reference genes for gene expression analysis in the brown planthopper, Nilaparvata lugens (Hemiptera: Delphacidae) using reverse-transcription quantitative PCR. PLoS ONE 9:e86503. https://doi.org/10.1371/journal.pone.0086503
Zhang S, Zeng Y, Yi X, Zhang Y (2016) Selection of suitable reference genes for quantitative RT-PCR normalization in the halophyte Halostachys caspica under salt and drought stress. Sci Rep 6:30363. https://doi.org/10.1038/srep30363
Zhu X, Li X, Chen W, Chen J, Lu W, Chen L, Fu D (2012) Evaluation of new reference genes in papaya for accurate transcript normalization under different experimental conditions. PLoS ONE 7:e44405. https://doi.org/10.1371/journal.pone.0044405
Funding
This work was funded by the National Natural Science Foundation of China (No. 31260181).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Phenotypic characteristics of one-year old seedlings of H. ammodendron. (a) General view of an untreated one-year old seedling growing in a tube filled with sand. (b) Assimilation branches separated from an untreated one-year old seedlings (Bar = 2 cm) (JPEG 218 kb)
Supplementary Fig. 2
Amplification of full length coding domains of ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10. M:DL 2000 DNA marker. Unit: bp (JPEG 722 kb)
Supplementary Fig. 3
Specificity of qRT-PCR amplicons. (a) 1.8% agarose gel electrophoresis showing amplification of a single product at the expected size for each reference gene. M represents 100 bp and 250 bp DNA Ladder. Lane 1-8 indicate ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10, respectively. (b) Dissociation curves with single peak were generated from all amplicons (JPEG 1060 kb)
Supplementary Table 1
Primers for cloning full length coding domains (ACT1, 18S, TUA5, ACT7, EF-1α, GAPDH, UBC18 and UBQ10) and qRT-PCR (FT-5, FT-9, DREB2A and DREB2C) (DOCX 39 kb)
Rights and permissions
About this article
Cite this article
Wang, B., Du, H., Yao, Z. et al. Validation of reference genes for accurate normalization of gene expression with quantitative real-time PCR in Haloxylon ammodendron under different abiotic stresses. Physiol Mol Biol Plants 24, 455–463 (2018). https://doi.org/10.1007/s12298-018-0520-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-018-0520-9