Abstract
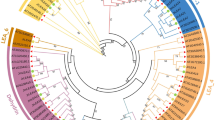
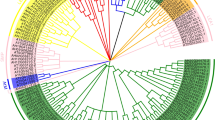
Late embryogenesis abundant (LEA) proteins are large and diverse group of polypeptides which were first identified during seed dehydration and then in vegetative plant tissues during different stress responses. Now, gene family members of LEA proteins have been detected in various organisms. However, there is no report for this protein family in watermelon and melon until this study. A total of 73 LEA genes from watermelon (ClLEA) and 61 LEA genes from melon (CmLEA) were identified in this comprehensive study. They were classified into four and three distinct clusters in watermelon and melon, respectively. There was a correlation between gene structure and motif composition among each LEA groups. Segmental duplication played an important role for LEA gene expansion in watermelon. Maximum gene ontology of LEA genes was observed with poplar LEA genes. For evaluation of tissue specific expression patterns of ClLEA and CmLEA genes, publicly available RNA-seq data were analyzed. The expression analysis of selected LEA genes in root and leaf tissues of drought-stressed watermelon and melon were examined using qRT-PCR. Among them, ClLEA-12-17-46 genes were quickly induced after drought application. Therefore, they might be considered as early response genes for water limitation conditions in watermelon. In addition, CmLEA-42-43 genes were found to be up-regulated in both tissues of melon under drought stress. Our results can open up new frontiers about understanding of functions of these important family members under normal developmental stages and stress conditions by bioinformatics and transcriptomic approaches.






Similar content being viewed by others
References
Altunoglu YC, Baloglu P, Yer EN, Pekol S, Baloglu MC (2016) Identification and expression analysis of LEA gene family members in cucumber genome. Plant Growth Regul. doi:10.1007/s10725-016-0160-4
Ambros V, Chen X (2007) The regulation of genes and genomes by small RNAs. Development 134(9):1635–1641
Arteaga-Vázquez M, Caballero-Pérez J, Vielle-Calzada JP (2006) A family of microRNAs present in plants and animals. Plant Cell 18:3355–3369
Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In: Proceedings of the second international conference on intelligent systems for molecular biology, AAAI Press, Menlo Park, CA, pp 28–36
Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014) Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 9(4):e96014. doi:10.1371/journal.pone.0096014
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2):281–297
Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Alejandra A (2008) The enigmatic LEA proteins and other hydrophilins. Plant Physiol 148:6–24
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The protein data bank. Nucleic Acids Res 28:235–242
Cao J, Li X (2015) Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 241(3):757–772
Cao J, Shi F (2012) Evolution of the RALF gene family in plants: gene duplication and selection patterns. Evol Bioinform 8:271–292
Charfeddine S, Saïdi MN, Charfeddine M, Gargouri-Bouzid R (2015) Genome wide identification and expression profiling of the late embryogenesis abundant genes in potato with emphasis on dehydrins. Mol Biol Rep 42(7):1163–1174
Conesa A, Götz S (2008) Blast2GO: a comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008:619832
Dai X, Zhao PX (2011) psRNATarget: a plant small RNA target analysis server. Nucleic Acids Res. doi:10.1093/nar/GKR319
Dalal M, Tayal D, Chinnusamy V, Bansal KC (2009) Abiotic stress and ABA-inducible group 4 LEA from Brassica napus plays a key role in salt and drought tolerance. J Biotechnol 139(2):137–145. doi:10.1016/j.jbiotec.2008.09.014
Denekamp NY, Reinhardt R, Kube M, Lubzens E (2010) Late embryogenesis abundant (LEA) proteins in nondesiccated, encysted, and diapausing embryos of rotifers. Biol Reprod 82(4):714–724. doi:10.1095/biolreprod.109.081091
Du D, Zhang Q, Cheng T et al (2013) Genome-wide identification and analysis of late embryogenesis abundant (LEA) genes in Prunus mume. Mol Biol Rep 40(2):1937–1946. doi:10.1007/s11033-012-2250-3
Duan J, Cai W (2012) OsLEA3-2, an abiotic stress induced gene of rice plays a key role in salt and drought tolerance. PLoS ONE 7:e45117
Dure LI, Galau GA (1981) Developmental biochemistry of cotton seed embryogenesis and germination XIII. Regulation of biosynthesis of principal storage proteins. Plant Physiol 68(1):187–194
Dure L, Crouch M, Harada JJ, Ho T, Mundy J, Quatrano RS, Thomas TL, Sung ZR (1989) Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol Biol 12:475–486
Eichinger L, Pachebat JA, Glockner G, Rajandream MA, Sucgang R, Berriman M (2005) The genome of the social amoeba Dictyostelium discoideum. Nature 435:43–57
Eulgem T, Rushton PJ, Robatzek S, Somssich IE (2000) The WRKY superfamily of plant transcription factors. Trends Plant Sci 5:199–206
Filiz E, Ozyigit II, Tombuloglu H, Koc I (2013) In silico comparative analysis of LEA (late embryogenesis abundant) proteins in Brachypodium distachyon L. Plant Omics 6(6):433
Flaus A et al (2006) Identification of multiple distinct SNF2 subfamilies with conserved structural motifs. Nucleic Acids Res 43:2887–2905
Gal TZ, Glazer I, Koltai H (2004) An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 577:21–26
Galau GA, Hughes DW, Dure L (1986) Abscisic acid induction of cloned cotton late embryogenesis abundant (LEA) messenger RNAs. Plant Mol Biol 7:155–170
Garcia-Mas J, Benjak A, Sanseverino W, Bourgeois M, Mir G, González VM, Alioto T et al (2012) The genome of melon (Cucumis melo L.). Proc Natl Acad Sci 109(29):11872–11877
Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, Rokshar DS (2012) Phytozome: a 14 comparative platform for green plant genomics. Nucleic Acids Res 40:D1178–D1186
Goyal K, Walton LJ, Tunnacliffe A (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157
Grelet J, Benamar A, Teyssier E et al (2005) Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol 137:157–167
Guleria P, Yadav SK (2011) Identification of miR414 and expression analysis of conserved miRNAs from Stevia rebaudiana. Genom Proteom Bioinform 9(6):211–217
Guo Q et al (2007) Bioinformatic identification of microRNAs and their target genes from Solanum tuberosum expressed sequence tags. Chin Sci Bull 52:2380–2389
Guo S, Zhang J, Sun H, Salse J, Lucas WJ, Zhang H, Min J et al (2013) The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat Genet 45(1):51–58
Gurley WB (2000) HSP101: a key component for the acquisition of thermotolerance in plants. Plant Cell 12:457–460
Hara M, Terashima S, Kuboi T (2001) Characterization and cryoprotective activity of cold-responsive dehydrin from Citrus unshiu. J Plant Physiol 158:1333–1339
He S, Tan L, Hu Z, Chen G, Wang G, Hu T (2012) Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol Genet Genom 287:39–54
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Agric Exp Stn Circ 347:1–32
Hunault G, Jaspard E (2010) LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genom 11:221. doi:10.1186/1471-2164-11-221
Hundertmark M, Hincha DK (2008) LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genom 9:118. doi:10.1186/1471-2164-9-118
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F, bZIP Research Group (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111
Jeon OS, Kim CS, Lee SP, Kang SK, Kim CM, Kang BG, Hur Y, Kim IJ (2006) Fruit ripening-related expression of a gene encoding group 5 late embryogenesis abundant protein in Citrus. J Plant Biol 49:403–408
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10(6):845–858
Kikawada T, Nakahara H, Kanamori Y, Iwata K, Watanabe M, McGee B, Tunnacli A, Okuda T (2006) Dehydration induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun 348:56–61
Kosová K, Vítámvás P, Prášil IT (2007) The role of dehydrins in plant response to cold. Biol Plant 51(4):601–617
Kumar R, Wehner TC (2011) Breeding for high yield—a review. Cucurbit Genet Coop Rpt 33/34:41–42
Lan T, Gao J, Zeng QY (2013) Genome-wide analysis of the LEA (late embryogenesis abundant) protein gene family in Populus trichocarpa. Tree Genet Genomes 9(1):253–264
Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. doi:10.1093/nar/gkr931
Li X, Cao J (2016) Late Embryogenesis Abundant (LEA) gene family in maize: identification, evolution, and expression profiles. Plant Mol Biol Report 34(1):15–28
Li L, Xu HL, Yang XL, Li YX, Hu YK (2011) Genome-wide identification, classification and expression analysis of LEA gene family in soybean. Sci Agric Sin 44(19):3945–3954
Liu Y, Zheng Y (2005) PM2, a group3 LEA protein from soybean, and its22-merrepeatingregion confersal tolerance in Escherichia coli. Biochem Biophys Res Commun 331:325–332
Liu SQ, Xu L, Jia ZQ, Xu Y, Yang Q, Fei ZJ, Lu XY, Chen HM, Huang SW (2004) Genetic association of ethylene-insensitive-3-like sequence with the sex-determining M locus in cucumber (Cucumis sativus L.). Theor Appl Genet 117:927–933
Liu Y, Zheng Y, Zhang Y, Wang W, Li R (2010) Soybean PM2 protein (LEA3) confers the tolerance of Escherichia coli and stabilization of enzyme activity under diverse stresses. Curr Microbiol 60:373–378
Livak JK, Schmittgen TD (2001) Analysis of relative gene expression data using real time quantitative PCR and the 2−ΔΔCt method. Methods 25:402–408
Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155
Manfre A, Lanni LM, Marcotte WR (2006) The Arabidopsis group 1 late embryogenesis abundant protein ATEM6 is required for normal seed development. Plant Physiol 140:140–149
Marunde MR, Samarajeewa DA, Anderson J, Li S, Hand SC, Menze MA (2013) Improved tolerance to salt and water stress in Drosophila melanogaster cells conferred by late embryogenesis abundant protein. J Insect Physiol 59(4):377–386
Mehan MR, Freimer NB, Ophoff RA (2004) A genome-wide survey of segmental duplications that mediate common human genetic variation of chromosomal architecture. Hum Genom 1(5):335–344
Naot D, Ben-Hayyim G, Eshdat Y, Holland D (1995) Drought, heat and salt stress induce the expression of a citrus homologue of an atypical late-embryogenesis Lea5 gene. Plant Mol Biol 27:619–622
Olvera-Carrillo Y, Campos F, Luis Reyes J, Garciarrubio A, Covarrubias AA (2010) Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol 154(1):373–390. doi:10.1104/pp.110.158964
Pani A, Mahapatra RK (2013) Computational identification of microRNAs and their targets in Catharanthus roseus expressed sequence tags. Genom Data 1:2–6
Pedrosa AM, Martins CDPS, Gonçalve SLP, Costa MGC (2015) Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 10(12):e0145785
Puhakainen T, Hess MW, Mäkelä P, Svensson J, Heino P, Palva ET (2004) Overexpression of multiple dehydrin genes enhances tolerance to freezing stress in Arabidopsis. Plant Mol Biol 54:743–753
Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N et al (2005) InterProScan: protein domains identifier. Nucleic Acids Res 33:W116–W120
Reyes JL, Campos F, Wei H, Arora R, Yang Y, Karlson DT, Covarrubias AA (2008) Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ 31:1781–1790
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Shih MD, Lin SC, Hsieh JS, Tsou CH, Chow TY, Lin TP, Hsing YI (2004) Gene cloning and characterization of a soybean (Glycine max L.) LEA protein, GmPM16. Plant Mol Biol 56(5):689–703
Singh S, Cornilescu CC, Tyler RC, Cornilescu G, Tonelli M, Lee MS, Markley JL (2005) Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stressrelated protein. Protein Sci 14(10):2601–2609
Suo J et al (2003) Identification of GhMYB109 encoding a R2R3MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochim Biophys Acta 1630:25–34
Suyama M, Torrents D, Bork P (2006) PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34:W609–W612
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729. doi:10.1093/molbev/mst197
Tang H, Bowers JE, Wang X, Ming R, Alam M et al (2008) Synteny and collinearity in plant genomes. Science 320:486–488
Tolleter D, Hincha DK, Macherel D (2010) A mitochondrial late embryogenesis abundant protein stabilizes model membranes in the dry state. Biochim Biophys Acta 1798:1926–1933
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang XS, Zhu HB, Jin GL, Liu HL, Wu WR, Zhu J (2007) Genome-scale identification and analysis of LEA genes in rice (Oryza sativa L.). Plant Sci 172(2):414–420
Wolkers WF, McCready S, Brandt WF, Lindsey GG, Hoekstra FA (2001) Isolation and characterization of a D-7 LEA protein from pollen that stabilizes glasses in vitro. Biochim Biophys Acta 1544(1–2):196–206
Yang Z, Gu S, Wang X, Li W, Tang Z et al (2008) Molecular evolution of the cpp2 like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol 67:266–277
Zhang Y (2005) miRU: an automated plant miRNA target prediction server. Nucleic Acids Res 33:W701–W704
Zhang WS et al (2003) Interaction of wheat high-mobility-group proteins with four-way-junction DNA and characterization of the structure and expression of HMGA gene. Arch Biochem Biophys 409:357–366
Zhou L, Liu Y, Liu Z, Kong D, Duan M, Luo L (2010) Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J Exp Bot 61(15):4157–4168
Acknowledgements
This work was financially supported by The Scientific and Technological Research Council of Turkey (TUBITAK) with Grant Number 114O761.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Presentation of tissue specific expression of ClLEA genes by heatmap from different tissues of watermelon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 7607 kb)
Supplementary Fig. 2
Tissue specific expression patterns of CmLEA genes by heatmap from different tissues of melon. Expression levels of genes were presented by colors from low (green) to high (red) expression pattern. (TIFF 4328 kb)
Supplementary Fig. 3
A total of 10 different motif clades with different colored boxes in melon (A) and watermelon (B) LEA proteins by MEME software. (TIFF 4075 kb)
Supplementary Fig. 4
Phylogenetic relationships between melon, watermelon and cucumber LEA proteins. (TIFF 2718 kb)
Supplementary Fig. 5
Comparative physical mapping of ClLEA genes with Arabidopsis, poplar, soybean, potato, grape and maize LEA genes. (TIFF 4326 kb)
Rights and permissions
About this article
Cite this article
Celik Altunoglu, Y., Baloglu, M.C., Baloglu, P. et al. Genome-wide identification and comparative expression analysis of LEA genes in watermelon and melon genomes. Physiol Mol Biol Plants 23, 5–21 (2017). https://doi.org/10.1007/s12298-016-0405-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-016-0405-8