Abstract
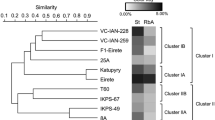
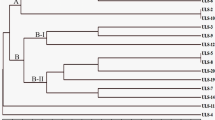
In the present study, molecular (DAMD and ISSR) and chemical (α and β-asarone contents) markers were used to characterize the A. calamus genotypes procured from different parts of India. The cumulative analysis carried out for both DAMD and ISSR markers revealed 24.71 % polymorphism across all genotypes of A. calamus. The clustering patterns of the genotypes in the UPGMA tree showed that the genotypes are diverse, and did not show any specific correlation with their geographical provenances, reflecting the low level of genetic diversity and a high genetic differentiation among the genotypes from the same localities. All the 27 genotypes of A. calamus were also analyzed for α and β-asarone contents, and percentage of essential oil. The genotype (Ac13) from Kullu (Himachal Pradesh) showed maximum (9.5 %) percentage of oil, whereas corresponding minimum (2.8 %) was obtained from the genotypes from Pangthang (Sikkim). Similarly, the highest α and β-asarone contents (16.82 % and 92.12 %) were obtained from genotypes from Renuka (Himachal Pradesh) and Udhampur (Jammu & Kashmir), while lowest α and β-asarone contents (0.83 % and 65.96 %) resulted from Auranwa (Uttar Pradesh) and Pangthang (Sikkim) genotypes, respectively. A. calamus harbours tremendous economic value, and it is therefore, important to identify the genotypes with low α and β-asarone contents for its commercial utilization. Further, this study will help in evaluation and documentation of a large number of diverse genotypes for their value traits.

Similar content being viewed by others
References
Abdul Kareem VK, Rajasekharan PE, Ravish BS, Mini S, Sane A, Kumar TV (2012) Analysis of genetic diversity in Acorus calamus populations in South and North East India using ISSR markers. Biochem Syst Ecol 40:156–161
Ahlawat A, Katoch M, Ram G, Ahuja A (2010) Genetic diversity in Acorus calamus L. as revealed by RAPD markers and its relationship with b-asarone content and ploidy level. Sci Hortic 124:294–297
Bertea CM, Azzolin CMM, Bossi S, Doglia G, Maffei ME (2005) Identification of an EcoRI restriction site for a rapid and precise determination of b-asarone-free Acorus calamus cytotypes. Phytochemistry 66:507–514
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Chang PS (2010) Flora of China. 23:1–2. Available from: http://www.efloras.org
Ginwal HS, Mittal N, Barthwal S (2009) Development and characterization of polymorphic chloroplast microsatellite markers in sweet flag (Acorus calamus L.). Indian J Genet 69:256–259
Heath DD, Iwama GK, Devlin RH (1993) PCR primed with the VNTR core sequences yields species specific patterns and hypervariable probes. Nucleic Acids Res 21:5782–5785
Janki Amal EK, Sobti SN, Handa KL (1964) The interrelationship between polyploidy, altitude and chemical composition in Acorus calamus. Curr Sci 33:500
Karthikeyan S, Jain SK, Nayar MP, Sanjappa M (1989) Florae Indicae Enumeratio Monocotyledonae. Botanical Survey of India, Calcutta
Krahulcova A (2003) Chromosome numbers in selected monocotyledons. Perslia Praha 75:97–113
Liao LC, Hsiao JY (1998) Relationship between population genetic structure and riparian habitat as revealed by RAPD analysis of the rheophyte Acorus gramineus Soland. (Araceae) in Taiwan. Mol Ecol 7:1275–1281
Mcgaw LJ, Jager AK, Staden J (2002) Isolation of b-asarone, an antibacterial and anthelmintic compound, from Acorus calamus in South Africa. S Afr J Bot 68:31–35
Mehrotra S, Mishra KP, Maurya R, Srimal RC, Yadav VS, Pandey R, Singh VK (2003) Anticellular and immunosuppressive properties of ethanolic extract of Acorus calamus rhizome. Int Immunopharmacol 3:53–61
Mittal N, Ginwal HS, Varshney VK (2009) Pharmaceutical and biotechnological potential of Acorus calamus Linn.: an indigenous highly valued medicinal plant species. Pharmacogn Rev 3:93–103
Ogra RK, Mahapatra P, Sharma UK, Sharma M, Sinha AK, Ahuja PS (2009) Indian calamus (Acorus calamus L.): not a tetraploid. Curr Sci 97:1644–1647
Page RDM (2001) TreeView (Win32), Ver. 1.6.5. Available from: http://taxonomy.zoology.gla.ac.uk/rod/treeview.html
Pai A, McCarthy BC (2005) Variation in shoot density and rhizome biomass of Acorus calamus L. with respect to environment. Castanea 70:263–275
Pavlicek A, Hrda S, Flegr J (1999) Free Tree - Freeware program for construction of phylogenetic trees on the basis of distance data and bootstrapping / jackknife analysis of the tree robustness. Application in the RAPD analysis of the genus Frenkelia. Folia Biol (Praha) 45:97–99, Available from: http://www.natur.cuni.cz/~flegr/programs/freetree.htm
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Provost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Raina VK, Srivastava SK, Syamasunder KV (2003) Essential oil composition of Acorus calamus L. from the lower region of the Himalayas. Flav Frag J 18:18–20
Ravindran PN, Balachandran I (2004) Underutilized medicinal spices (1). Spice India 17:1–14
Riaz M, Qamar S, Chaudhary FM (1995) Chemistry of the medicinal plants of the genus Acorus (family Araceae). Hamdard Med 38:50–62
Rost LCM, Bos R (1979) Biosystematic investigation with Acorus L. 3 Communication. Constituents of essential oils. Planta Med 36:350–361
Todorova MN, Ognyanov IV, Shatar S (1995) Chemical composition of essential oil from Mongolian Acorus calamus L. rhizomes. J Essent Oil Res 7:191–193
Zhou Z, Bebeli PJ, Somers DJ, Gustafson JP (1997) Direct amplification of minisatellite-region DNA with VNTR core sequences in the genus Oryza. Theor Appl Genet 95:942–949
Acknowledgements
The authors are thankful to the Director, CSIR-National Botanical Research Institute, Lucknow for facilities and encouragements. The authors (MMP, SKS and AKSR) are also thankful to the NAIP (ICAR, New Delhi) for financial support.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rana, T.S., Mahar, K.S., Pandey, M.M. et al. Molecular and chemical profiling of ‘sweet flag’ (Acorus calamus L.) germplasm from India. Physiol Mol Biol Plants 19, 231–237 (2013). https://doi.org/10.1007/s12298-013-0164-8
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-013-0164-8