Abstract
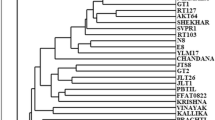
In this study, the genetic relatedness of 82 walnut genotypes adapted to the North Western Himalayan region of Jammu and Kashmir, India was analyzed by combination of 13 SSR and 20 RAPD primers. A high level of genetic diversity was observed within populations with the number of alleles per locus ranging from one to five in case of SSR primers and two to six in case of RAPD primers, the proportion of polymorphic loci was 100 %, and similarity ranged from 12 % to 79 % with an average of 49 %. Dendrogram showed that all the accessions formed four main clusters with various degree of sub-clustering within the clusters. These results have implications for walnut breeding and conservation.





Similar content being viewed by others
References
Ana CR, Paul CA, Myriam CD, Maria MM, Merideth WB, Carlos I, Joe T (2000) Cross-species amplification of cassava (Manihot esculenta) (Euphorbiaceae) microsatellites: Allelic polymorphism and degree of relationship. Am J Bot 87:1647–1655
Anderson JA, Churchill JE, Autrique SD, Tanksley S, Sorrells ME (1993) Optimizing parental selection for genetic linkage maps. Genome 36:181–188
Bayazit S, Kazan K, Gulbitti S, Çevik V, Ayanoglu H, Ergul A (2007) AFLP analysis of genetic diversity in low chill requiring walnut (Juglans regia L.) genotypes from Hatay, Turkey. Sci Hort 111:394–398
Busov VB, Rink G, Woeste KE (2002) Allozyme variation and mating system of black walnut (Juglans nigra L.) in the central hardwood region of the United States. For Genet 9:319–327
Carriero F, Fontanazza G, Cellini F, Giorio G (2002) Identification of simple sequenze repeats (SSRs) in olive (Olea europaea L.). Theor Appl Genet 104:301–307
Casas AM, Igartua E, Balaguer G, Moreno MA (1999) Genetic diversity of Prunus rootstocks analyzed by RAPD markers. Euphytica 110:139–149
Ciarmiello LF, Piccirillo P, Pontecorvo G, Luca AD, Kafantaris I, Woodrow P (2011) A PCR based SNPs marker for specific characterization of English walnut (Juglans regia L.) cultivars. Mol Biol Rep 38:1237–1249
Dangl GS, Woeste K, Aradhya MK, Koehmstedt A, Simon C, Potter D, Leslie CA, McGranahan G (2005) Characterization of 14 microsatellite markers for genetic analysis and cultivar identification of walnut. J Am Soc Hort Sci 130:348–354
Ebrahimi A, Fatahi R, Zamani Z (2011) Analysis of genetic diversity among some Persian walnut genotypes (Juglans regia L.) using morphological traits and SSRs markers Scientia. Sci Hortic 130:146–151
Fjellstrom RG, Parfitt DE (1995) Phylogenetic analysis and evolution of the genus Juglans (Juglandaceae) as determined from nuclear genome RFLPs. Plant Syst Evol 197:19–32
Fornari B, Malvolti ME, Taurchini D, Fineschi S, Beritognolo I, McCaglia E, Cannata F (2001) Isozyme and organellar DNA analysis of genetic diversity in natural/ naturalized European and Asiatic walnut (Juglans regia) populations. Acta Hort 544:167–178
Foroni I, Rao R, Woeste K, Gallitelli M (2005) Characterization of Juglans regia L. through SSR markers and evaluation of genetic relationships among cultivars and the ‘Sorrento’ landrace. J Hort Sci Biotech 80:49–53
Foroni I, Woeste K, Monti LM, Rao R (2007) Identification of ‘Sorrento’ walnut using simple sequence repeats (SSRs). Genet Res Crop Evol 54:1081–109
Francesca PI, Damfil D, Raica P, Petricele IV, Sisea C, Vas C, Botos B, Bodea M, Botu M (2010) Assessment of the genetic variability among some Juglans cultivars from the Romanian National Collection at S.C.D. P. Vâlcea using RAPD markers. Rom Biotechnol Lett 15:41–49
Galderisi U, Cipollaro M, Bernardo DI, Masi DE, Galano G, Cascino A (1999) Identification of Hazelnut (Corylus avellana) cultivars by RAPD analysis. Plant Cell Rep 18:652–655
Goulao L, Cabrita L, Oliveira CM, Leitao JM (2001) Comparing RAPD and AFLP analysis in discrimination and estimation of genetic similarities among apple (Malus domestica Borkh.) cultivars. Euphytica 199:259–270
Gunn BF, Aradhya M, Salick JM, Miller AJ, Yongping Y, Lin L, Xian H (2010) Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): species distinctions, human impacts, and the conservation of agrobiodiversity in Yunnan, China. Am J Bot 97:660–671
Gupta PK, Varshney RK (2000) The development and use of microsatellite markers for genetic analysis and plant breeding with emphasis on bread wheat. Euphytica 113:163–185
Hamza S, Hamida WB, Rebai A, Harrabi M (2004) SSR based genetic diversity among Tunisian Barley and relationship with morphological traits. Euphytica 135:107–118
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull Soc Vaud Sci Nat 44:223–270
Kafkas S, Ozkan H, Sutyemez M (2005) DNA polymorphism and assessment of genetic relationships in walnut genotypes based on AFLP and SAMPL markers. J Am Soc Hort Sci 130:585–590
Karimi R, Ershadi A, Vahdati K, Woeste K (2010) Molecular charac- terization of Persian walnut populations in Iran with microsatel-lite markers. Hort Sci 45:1403–1406
Li Z, Lanying Z, Qianwen X (2007) Identification of RAPD markers linked to thickness gene of shuck in walnut. Adv Biol Res 1:137–140
Litt M, Luly JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 44:397–401
Manning WE (1978) The classification with in Juglandaceae. Ann MO Bot Gard 65:1058–1087
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Mir JI, Karmakar P, Chattopadhyay S, Chaudhury SK, Ghosh SK, Roy A (2008) A Grouping of jute germplasm based on trait for fibre fineness through RAPD and SSR profiling and identification of contrasting parents—a prerequisite for mapping population development. In: Proceedings of International symposium on Jute and Allied Fibres Production, Utilization and Marketing, Kolkata, India. pp 26
Nicese FP, Hormaza JI, McGranahan GH (1998) Molecular characterization and genetic relatedness among walnut (Juglans regia L.) genotypes based on RAPD markers. Euphytica 101:199–206
Ninot A, Aleta N (2003) Identification and genetic relationship of Persian walnut genotypes using isozyme markers. J Am Pomol Soc 57:106–114
Pollegioni P, Woeste K, Mugnozza GS, Malvolt ME (2009) Retrospective identification of hybridogenic walnut plants by SSR fingerprinting and parentage analysis. Mol Breed 24:321–335
Potter D, Fangyou G, Aiello G, Leslie G, McGranahan G (2002) Intersimple sequences repeat markers for fingerprinting and determining genetic relationships of walnut (Juglans regia) cultivars. J Am Soc Hort Sci 127:75–81
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalsky A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98:107–112
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Robert HW (1930) Meiosis of Microsporogenesis in the Juglandaceae. Am J Bot 17:863–869
Robichaud RL, Glaubitz JC, Rhodes OE, Woeste K (2006) A robust set of black walnut microsatellites for parentage and clonal identification. New Forests 32:179–196
Solar A, Podjavorsek A, Stamar F (2005) Phenotypic and genotypic diversity of European chestnut (Castanea sativa Mill.) in Slovenia—opportunity for genetic improvement. Gen Res Crop Evol 52:381–394
Streiff R, Labbe T, Bacilieri R (1998) Within population genetic structure in Quercus robur L. and Quercus petraea (Matt.) Liebl. Assessed with isozymes and microsatellites. Mol Ecol 7:317–328
Victory ER, Glaubitz JC, Rhodes OE, Woeste KE (2006) Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93:118–126
Vyas D, Sharma SK, Sharma DR (2003) Genetic structure of walnut genotypes using leaf isozymes as variability measure. Sci Hort 97:141–152
Weising K, Nybom H, Wolff K, Meyer W (1995) DNA fingerprinting in plants and fungi. CRC Press Inc, Boca Raton, Florida
Woeste K, Burns R, Rhodes O, Michler C (2002) Thirty polymorphic nuclear microsatellite loci from black walnut. J Hered 93:58–60
Yan-Min W, Ying L, Feng-Xiang D, Sheng-Ke X (2000) Study on different ecological types of Chinese walnut (Juglans regia) using RAPD markers. J Beij For Univ 22:23–27
Yap IP, Nelson RJ (1996) Winboot: A program for perform- ing bootstrap analysis of binary data to determine the confidemnce level. IRRI Discuss Paper Ser. 14, IRRI, Manila, Philippines
Zenelli G, Kola H, Dida M (2005) Phenotypic variation in native walnut populations of Northern Albania. Sci Hort 105:91–100
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ahmed, N., Mir, J.I., Mir, R.R. et al. SSR and RAPD analysis of genetic diversity in walnut (Juglans regia L.) genotypes from Jammu and Kashmir, India. Physiol Mol Biol Plants 18, 149–160 (2012). https://doi.org/10.1007/s12298-012-0104-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-012-0104-z