Abstract
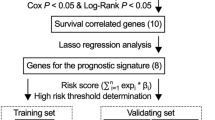
Although breast cancer (BC) has a low mortality rate relative to other cancers, it prominently affects the survival of patients with human epidermal growth factor receptor-2 (HER2 +) BC due to its high recurrence rate. By far, it has been found that autophagy can affect various tumor occurrence and development, as well as patients’ prognosis. HER2 + BC patient samples and autophagy-related genes (ARGs) were acquired from a public database, least absolute shrinkage and selection operator (LASSO) and Cox analyses (including univariate and multivariate analyses) were utilized to construct a 9-ARGs model, which was verified by using HER2 + BC patient samples in The Cancer Genome Atlas (TCGA) dataset. Sample risk score was worked out based on characteristic genes, and prominent differences in overall survival were tracked down between high- and low-risk groups. Predictive ability of the model was validated by drawing receiver operating characteristic (ROC) curves and then calculating the area under the curves (AUC) value. Results showed good accuracy and prediction ability of the model in both validation set and training set. For the purpose of facilitating model application in clinical practice, we constructed a nomogram combing clinical factors and risk scores to evaluate 1-year, 3-year and 5-year survival of HER2 + BC patients. In addition, we assessed the correlation of risk score with tumor mutational burden and tumor immune infiltration. Results exhibited that in a high-risk group, tumor mutation was relatively high, while tumor immune infiltration was relatively poor. Overall, based on ARGs, the prognostic signature in this study can tellingly evaluate prognoses of HER2 + BC patients and provide a reference for clinicians.







Similar content being viewed by others
Data availability
Data sharing is not applicable to this article as no datasets were generated or analysed during the current study.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49.
Cronin KA, Harlan LC, Dodd KW, Abrams JS, Ballard-Barbash R. Population-based estimate of the prevalence of HER-2 positive breast cancer tumors for early stage patients in the US. Cancer Invest. 2010;28(9):963–8.
Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235(4785):177–82.
Seshadri R, Firgaira FA, Horsfall DJ, McCaul K, Setlur V, Kitchen P. Clinical significance of HER-2/neu oncogene amplification in primary breast cancer. The South Australian breast cancer study group. J Clin Oncol. 1993;11(10):1936–42.
Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, Kimmelman A, Kumar S, Levine B, Maiuri MC, Martin SJ, Penninger J, Piacentini M, Rubinsztein DC, Simon HU, Simonsen A, Thorburn AM, Velasco G, Ryan KM, Kroemer G. Autophagy in malignant transformation and cancer progression. EMBO J. 2015;34(7):856–80.
Smit L, Berns K, Spence K, Ryder WD, Zeps N, Madiredjo M, Beijersbergen R, Bernards R, Clarke RB. An integrated genomic approach identifies that the PI3K/AKT/FOXO pathway is involved in breast cancer tumor initiation. Oncotarget. 2016;7(3):2596–610.
Schaffner I, Minakaki G, Khan MA, Balta EA, Schlotzer-Schrehardt U, Schwarz TJ, Beckervordersandforth R, Winner B, Webb AE, DePinho RA, Paik J, Wurst W, Klucken J, Lie DC. FoxO function is essential for maintenance of autophagic flux and neuronal morphogenesis in adult neurogenesis. Neuron. 2018;99(6):1188–12036.
Audesse AJ, Dhakal S, Hassell LA, Gardell Z, Nemtsova Y, Webb AE. FOXO3 directly regulates an autophagy network to functionally regulate proteostasis in adult neural stem cells. PLoS Genet. 2019;15(4):e1008097.
Lin QG, Liu W, Mo YZ, Han J, Guo ZX, Zheng W, Wang JW, Zou XB, Li AH, Han F. Development of prognostic index based on autophagy-related genes analysis in breast cancer. Aging (Albany NY). 2020;12(2):1366–76.
Zhao S, Guan B, Mi Y, Shi D, Wei P, Gu Y, Cai S, Xu Y, Li X, Yan D, Huang M, Li D. LncRNA MIR17HG promotes colorectal cancer liver metastasis by mediating a glycolysis-associated positive feedback circuit. Oncogene. 2021;40(28):4709–24.
Vega-Rubin-de-Celis S, Zou Z, Fernandez AF, Ci B, Kim M, Xiao G, Xie Y, Levine B. Increased autophagy blocks HER2-mediated breast tumorigenesis. Proc Natl Acad Sci U S A. 2018;115(16):4176–81.
Therneau TM, Grambsch PM. Modeling survival data: extending the cox model. New York.: Springer; 2000.
Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33(1):1–22.
Blanche P, Dartigues JF, Jacqmin-Gadda H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat Med. 2013;32(30):5381–97.
Huang C, Liu Z, Xiao L, Xia Y, Huang J, Luo H, Zong Z, Zhu Z. Clinical significance of serum CA125, CA19-9, CA72-4, and fibrinogen-to-lymphocyte ratio in gastric cancer with peritoneal dissemination. Front Oncol. 2019;9:1159.
Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7.
Schettini F, Prat A. Dissecting the biological heterogeneity of HER2-positive breast cancer. Breast. 2021;59:339–50.
Tyutyunyk-Massey L, Gewirtz DA. Roles of autophagy in breast cancer treatment: target, bystander or benefactor. Semin Cancer Biol. 2020;66:155–62.
Chiu CF, Chin HK, Huang WJ, Bai LY, Huang HY, Weng JR. Induction of apoptosis and autophagy in breast cancer cells by a novel HDAC8 inhibitor. Biomolecules. 2019;9(12):824.
Ulasov IV, Borovjagin AV, Timashev P, Cristofanili M, Welch DR. KISS1 in breast cancer progression and autophagy. Cancer Metastasis Rev. 2019;38(3):493–506.
Li X, Jin F, Li Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J Cell Mol Med. 2021;25(1):4–14.
Storr SJ, Thompson N, Pu X, Zhang Y, Martin SG. Calpain in breast cancer: role in disease progression and treatment response. Pathobiology. 2015;82(3–4):133–41.
Storr SJ, Woolston CM, Barros FF, Green AR, Shehata M, Chan SY, Ellis IO, Martin SG. Calpain-1 expression is associated with relapse-free survival in breast cancer patients treated with trastuzumab following adjuvant chemotherapy. Int J Cancer. 2011;129(7):1773–80.
Al-Bahlani SM, Al-Rashdi RM, Kumar S, Al-Sinawi SS, Al-Bahri MA, Shalaby AA. Calpain-1 expression in triple-negative breast cancer: a potential prognostic factor independent of the proliferative/apoptotic index. Biomed Res Int. 2017;2017:9290425.
Yeerken D, Hong R, Wang Y, Gong Y, Liu R, Yang D, Li J, Fan J, Chen J, Zhang W, Zhan Q. PFKP is transcriptionally repressed by BRCA1/ZBRK1 and predicts prognosis in breast cancer. PLoS ONE. 2020;15(5):e0233750.
Farshchian M, Kivisaari A, Ala-Aho R, Riihila P, Kallajoki M, Grenman R, Peltonen J, Pihlajaniemi T, Heljasvaara R, Kahari VM. Serpin peptidase inhibitor clade A member 1 (SerpinA1) is a novel biomarker for progression of cutaneous squamous cell carcinoma. Am J Pathol. 2011;179(3):1110–9.
Tan XF, Wu SS, Li SP, Chen Z, Chen F. Alpha-1 antitrypsin is a potential biomarker for hepatitis B. Virol J. 2011;8:274.
Chan HJ, Li H, Liu Z, Yuan YC, Mortimer J, Chen S. SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget. 2015;6(28):25815–27.
Song Y, Lu M, Qiu H, Yin J, Luo K, Zhang Z, Jia X, Zheng G, Liu H, He Z. Activation of FOXO3a reverses 5-Fluorouracil resistance in human breast cancer cells. Exp Mol Pathol. 2018;105(1):57–62.
Wang S, Huo D, Ogundiran TO, Ojengbede O, Zheng W, Nathanson KL, Nemesure B, Ambs S, Olopade OI, Zheng Y. Association of breast cancer risk and the mTOR pathway in women of African ancestry in ‘The Root’ Consortium. Carcinogenesis. 2017;38(8):789–96.
Chen D, Zhong Q. A tethering coherent protein in autophagosome maturation. Autophagy. 2012;8(6):985–6.
Wu M, Chen B, Pan X, Su J. Prognostic value of autophagy-related proteins in human gastric cancer. Cancer Manag Res. 2020;12:13527–40.
Eissa S, Matboli M, Awad N, Kotb Y. Identification and validation of a novel autophagy gene expression signature for human bladder cancer patients. Tumour Biol. 2017;39(4):1010428317698360.
John AS, Rothman VL, Tuszynski GP. Thrombospondin-1 (TSP-1) stimulates expression of integrin alpha6 in human breast carcinoma cells: a downstream modulator of tsp-1-induced cellular adhesion. J Oncol. 2010;2010:645376.
Brooks DL, Schwab LP, Krutilina R, Parke DN, Sethuraman A, Hoogewijs D, Schorg A, Gotwald L, Fan M, Wenger RH, Seagroves TN. ITGA6 is directly regulated by hypoxia-inducible factors and enriches for cancer stem cell activity and invasion in metastatic breast cancer models. Mol Cancer. 2016;15:26.
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61.
Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA. Genomic correlates of immune-cell infiltrates in colorectal Carcinoma. Cell Rep. 2016;15(4):857–65.
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med. 2014;371(23):2189–99.
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA. Cancer immunology Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–8.
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA. Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science. 2015;350(6257):207–11.
Muraro E, Comaro E, Talamini R, Turchet E, Miolo G, Scalone S, Militello L, Lombardi D, Spazzapan S, Perin T, Massarut S, Crivellari D, Dolcetti R, Martorelli D. Improved Natural Killer cell activity and retained anti-tumor CD8(+) T cell responses contribute to the induction of a pathological complete response in HER2-positive breast cancer patients undergoing neoadjuvant chemotherapy. J Transl Med. 2015;13:204.
Garaud S, Buisseret L, Solinas C, Gu-Trantien C, de Wind A, Van den Eynden G, Naveaux C, Lodewyckx JN, Boisson A, Duvillier H, Craciun L, Ameye L, Veys I, Paesmans M, Larsimont D, Piccart-Gebhart M, Willard-Gallo K. Tumor infiltrating B-cells signal functional humoral immune responses in breast cancer. JCI Insight. 2019. https://doi.org/10.1172/jci.insight.129641.
Funding
This study was funded by the Public welfare project of Jinhua Science and Technology Plan, (2021–4-045).
Author information
Authors and Affiliations
Contributions
Conceptualization: FW. Data curation: FW. Formal analysis: LF. Methodology: LF. Software: BF. Validation: BF. Investigation: CF. Writing—original draft: FW. Writing—review & editing: LF.
Corresponding author
Ethics declarations
Conflict of interest
Author Fan Wang declares that he/she has no conflict of interest. Author Linghui Fang declares that he/she has no conflict of interest. Author Bifei Fu declares that he/she has no conflict of interest. Author Chen Fan declares that he/she has no conflict of interest.
Ethics approval and consent to participate
This article does not contain any studies with human participants performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Wang, F., Fang, L., Fu, B. et al. Construction of a prognostic risk assessment model for HER2 + breast cancer based on autophagy-related genes. Breast Cancer 30, 478–488 (2023). https://doi.org/10.1007/s12282-023-01440-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-023-01440-x