Abstract
Background
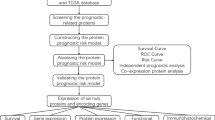
Proteomics profiles have enabled a systematic insight into the prognosis of cancer. This study aimed to establish a valuable protein-based risk signature to assess the prognosis and immune status in patients with breast cancer (BC).
Methods
Protein expression profile, RNA expression data, and clinical information were acquired from The Cancer Genome Atlas (TCGA). The whole cohort was randomly split into two cohorts, one for establishing the risk signature and the other for testing. Univariate Cox analysis and Least absolute shrinkage and selection operator (LASSO) Cox regression were utilized to construct the protein-based risk signature. All cohorts were divided into high- and low-risk groups, which were applied to investigate the clinical relevance, tumor microenvironment, and therapeutic response.
Results
The prognostic proteomics signature was established based on prognostic proteins, thus categorizing patients into low-risk and high-risk groups with different prognoses. A predictive nomogram was also developed to predict 1, 3, and 5-year survival possibility for BC patients, and the calibration curves confirmed the predictive significance of this signature. Afterward, the low-risk group displayed higher immune activities, immune checkpoint expression, and immunotherapeutic response. Moreover, GSEA analysis indicated that immune-associated pathways were rich in the low-risk group. Additionally, this prognostic signature demonstrated potential predict significance for chemotherapeutic agents.
Conclusion
This study established an effective prognostic proteomics signature with reliable predictive performance for survival, immune activity, and drug sensitivity. It might provide a novel perspective into the protein function in BC, and guide the individual treatment strategies for BC patients.







Similar content being viewed by others
Data availability
The data included in this research can be found in online databases.
References
Sun YS, Zhao Z, Yang ZN, Xu F, Lu HJ, Zhu ZY, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–97.
Barzaman K, Karami J, Zarei Z, Hosseinzadeh A, Kazemi MH, Moradi-Kalbolandi S, et al. Breast cancer: biology, biomarkers, and treatments. Int Immunopharmacol. 2020;84: 106535.
McDonald ES, Clark AS, Tchou J, Zhang P, Freedman GM. Clinical diagnosis and management of breast cancer. J Nucl Med. 2016;57(Suppl 1):9S-16S.
Rakha EA, Green AR. Molecular classification of breast cancer: what the pathologist needs to know. Pathology. 2017;49:111–9.
Cao X, Zhou Y, Mao F, Lin Y, Zhou X, Sun Q. Identification and characterization of three Siglec15-related immune and prognostic subtypes of breast-invasive cancer. Int Immunopharmacol. 2022;106: 108561.
Ma J, Chen C, Liu S, Ji J, Wu D, Huang P, et al. Identification of a five genes prognosis signature for triple-negative breast cancer using multi-omics methods and bioinformatics analysis. Cancer Gene Ther. 2022;29:1578–89.
Tang Y, Tian W, Xie J, Zou Y, Wang Z, Li N, et al. Prognosis and dissection of immunosuppressive microenvironment in breast cancer based on fatty acid metabolism-related signature. Front Immunol. 2022;13: 843515.
Yu H, Lv W, Tan Y, He X, Wu Y, Wu M, et al. Immunotherapy landscape analyses of necroptosis characteristics for breast cancer patients. J Transl Med. 2022;20:328.
Hermunen K, Soveri LM, Boisen MK, Mustonen HK, Dehlendorff C, Haglund CH, et al. Postoperative serum CA19-9, YKL-40, CRP and IL-6 in combination with CEA as prognostic markers for recurrence and survival in colorectal cancer. Acta Oncol. 2020;59:1416–23.
Hao C, Zhang G, Zhang L. Serum CEA levels in 49 different types of cancer and noncancer diseases. Prog Mol Biol Transl Sci. 2019;162:213–27.
Isgro MA, Bottoni P, Scatena R. Neuron-specific enolase as a biomarker: biochemical and clinical aspects. Adv Exp Med Biol. 2015;867:125–43.
Zhang M, Cheng S, Jin Y, Zhao Y, Wang Y. Roles of CA125 in diagnosis, prediction, and oncogenesis of ovarian cancer. Biochim Biophys Acta Rev Cancer. 2021;1875: 188503.
Van Poppel H, Albreht T, Basu P, Hogenhout R, Collen S, Roobol M. Serum PSA-based early detection of prostate cancer in Europe and globally: past, present and future. Nat Rev Urol. 2022;19:562–72.
Wang W, Xu X, Tian B, Wang Y, Du L, Sun T, et al. The diagnostic value of serum tumor markers CEA, CA19-9, CA125, CA15-3, and TPS in metastatic breast cancer. Clin Chim Acta. 2017;470:51–5.
Salmans ML, Zhao F, Andersen B. The estrogen-regulated anterior gradient 2 (AGR2) protein in breast cancer: a potential drug target and biomarker. Breast Cancer Res. 2013;15:204.
Pierga JY, Deneux L, Bonneton C, Vincent-Salomon A, Nos C, Anract P, et al. Prognostic value of cytokeratin 19 fragment (CYFRA 21–1) and cytokeratin-positive cells in bone marrow samples of breast cancer patients. Int J Biol Markers. 2004;19:23–31.
Chen C, Liu YQ, Qiu SX, Li Y, Yu NJ, Liu K, et al. Five metastasis-related mRNAs signature predicting the survival of patients with liver hepatocellular carcinoma. BMC Cancer. 2021;21:693.
Tibshirani R. The lasso method for variable selection in the Cox model. Stat Med. 1997;16:385–95.
Abdelhafez OH, Othman EM, Fahim JR, Desoukey SY, Pimentel-Elardo SM, Nodwell JR, et al. Metabolomics analysis and biological investigation of three Malvaceae plants. Phytochem Anal. 2020;31:204–14.
Goel MK, Khanna P, Kishore J. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res. 2010;1:274–8.
Huang R, Liao X, Li Q. Identification and validation of potential prognostic gene biomarkers for predicting survival in patients with acute myeloid leukemia. Onco Targets Ther. 2017;10:5243–54.
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–59.
Ma Q, Chen Y, Xiao F, Hao Y, Song Z, Zhang J, et al. A signature of estimate-stromal-immune score-based genes associated with the prognosis of lung adenocarcinoma. Transl Lung Cancer Res. 2021;10:1484–500.
Shen Y, Peng X, Shen C. Identification and validation of immune-related lncRNA prognostic signature for breast cancer. Genomics. 2020;112:2640–6.
Cheng Q, Chen X, Wu H, Du Y. Three hematologic/immune system-specific expressed genes are considered as the potential biomarkers for the diagnosis of early rheumatoid arthritis through bioinformatics analysis. J Transl Med. 2021;19:18.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50.
Prior FW, Clark K, Commean P, Freymann J, Jaffe C, Kirby J, et al. TCIA: An information resource to enable open science. Annu Int Conf IEEE Eng Med Biol Soc. 2013;2013:1282–5.
Yang W, Soares J, Greninger P, Edelman EJ, Lightfoot H, Forbes S, et al. Genomics of drug sensitivity in cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41:D955–61.
Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS ONE. 2014;9: e107468.
Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3.
Li X, Wang M, Li S, Chen Y, Wang M, Wu Z, et al. HIF-1-induced mitochondrial ribosome protein L52: a mechanism for breast cancer cellular adaptation and metastatic initiation in response to hypoxia. Theranostics. 2021;11:7337–59.
Qin J, Zhou Z, Chen W, Wang C, Zhang H, Ge G, et al. BAP1 promotes breast cancer cell proliferation and metastasis by deubiquitinating KLF5. Nat Commun. 2015;6:8471.
Lupu R, Cardillo M, Cho C, Harris L, Hijazi M, Perez C, et al. The significance of heregulin in breast cancer tumor progression and drug resistance. Breast Cancer Res Treat. 1996;38:57–66.
Alkarain A, Slingerland J. Deregulation of p27 by oncogenic signaling and its prognostic significance in breast cancer. Breast Cancer Res. 2004;6:13–21.
Truax AD, Thakkar M, Greer SF. Dysregulated recruitment of the histone methyltransferase EZH2 to the class II transactivator (CIITA) promoter IV in breast cancer cells. PLoS ONE. 2012;7: e36013.
Kabakov AE, Gabai VL. HSP70s in breast cancer: promoters of tumorigenesis and potential targets/tools for therapy. Cells. 2021;10:3446.
Du C, Wang Y, Zhang Y, Zhang J, Zhang L, Li J. LncRNA DLX6-AS1 Contributes to epithelial-mesenchymal transition and cisplatin resistance in triple-negative breast cancer via modulating mir-199b-5p/paxillin axis. Cell Transplant. 2020;29:963689720929983.
Funding
There is no funding to report.
Author information
Authors and Affiliations
Contributions
Conceptualization, XQ; writing–original draft pre-paration, XQ; writing–review and editing, XQ, CY and KW. All authors reviewed and approved the final version of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Qing, X., Yuan, C. & Wang, K. Characterization of protein-based risk signature to predict prognosis and evaluate the tumor immune environment in breast cancer. Breast Cancer 30, 424–435 (2023). https://doi.org/10.1007/s12282-023-01435-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-023-01435-8



