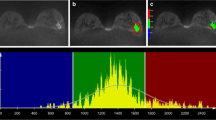
Abstract
Purpose
To investigate effective model composed of features from ultrafast dynamic contrast-enhanced magnetic resonance imaging (UF-MRI) for distinguishing low- from non-low-grade ductal carcinoma in situ (DCIS) lesions or DCIS lesions upgraded to invasive carcinoma (upgrade DCIS lesions) among lesions diagnosed as DCIS on pre-operative biopsy.
Materials and methods
Eighty-six consecutive women with 86 DCIS lesions diagnosed by biopsy underwent UF-MRI including pre- and 18 post-contrast ultrafast scans (temporal resolution of 3 s/phase). The last phase of UF-MRI was used to perform 3D segmentation. The time point at 6 s after the aorta started to enhance was used to obtain subtracted images. From the 3D segmentation and subtracted images, enhancement, shape, and texture features were calculated and compared between low- and non-low-grade or upgrade DCIS lesions using univariate analysis. Feature selection by least absolute shrinkage and selection operator (LASSO) algorithm and k-fold cross-validation were performed to evaluate the diagnostic performance.
Results
Surgical specimens revealed 16 low-grade DCIS lesions, 37 non-low-grade lesions and 33 upgrade DCIS lesions. In univariate analysis, five shape and seven texture features were significantly different between low- and non-low-grade lesions or upgrade DCIS lesions, whereas enhancement features were not. The six features including surface/volume ratio, irregularity, diff variance, uniformity, sum average, and variance were selected using LASSO algorism and the mean area under the receiver operating characteristic curve for training and validation folds were 0.88 and 0.88, respectively.
Conclusion
The model with shape and texture features of UF-MRI could effectively distinguish low- from non-low-grade or upgrade DCIS lesions.




Similar content being viewed by others
Abbreviations
- DCIS:
-
Ductal carcinoma in situ
- DCE-MRI:
-
Dynamic contrast-enhanced MRI
- UF-MRI:
-
Ultrafast DCE-MRI
- GLCM:
-
Gray level co-occurrence matrix
- ROI:
-
Region of interest
- IER:
-
Initial enhancement rate
- SER:
-
Signal enhancement ratio
- LASSO:
-
Least absolute shrinkage and selection operator
References
Silverstein MJ. Ductal carcinoma in situ of the breast. Annu Rev Med. 2000;51:17–32.
Sanders ME, Schuyler PA, Simpson JF, et al. Continued observation of the natural history of low-grade ductal carcinoma in situ reaffirms proclivity for local recurrence even after more than 30 years of follow-up. Mod Pathol. 2015;28:662–9.
Weaver DL, Rosenberg RD, Barlow WE, et al. Pathologic findings from the Breast Cancer Surveillance Consortium: population-based outcomes in women undergoing biopsy after screening mammography. Cancer. 2006;106:732–42.
Cserni G, Sejben A. Grading ductal carcinoma in situ (DCIS) of the breast—what’s wrong with It? Pathol Oncol Res. 2020;26:665–71.
Oseni TO, Smith BL, Lehman CD, et al. Do eligibility criteria for ductal carcinoma in situ (DCIS) active surveillance trials identify patients at low risk for upgrade to invasive carcinoma? Ann Surg Oncol. 2020;27:4459–65.
The ICSN DCIS Working Group, Ponti A, Ronco G, et al (2019) Low-grade screen-detected ductal carcinoma in situ progresses more slowly than high-grade lesions: evidence from an international multi-centre study. Breast Cancer Res. Treat 177:761–765
Maxwell AJ, Clements K, Hilton B, et al. Risk factors for the development of invasive cancer in unresected ductal carcinoma in situ. Eur J Surg Oncol. 2018;44:429–35.
Cserni G. Tumour histological grade may progress between primary and recurrent invasive mammary carcinoma. J Clin Pathol. 2002;55:293–7.
Wang S-Y, Shamliyan T, Virnig BA, et al. Tumor characteristics as predictors of local recurrence after treatment of ductal carcinoma in situ: a meta-analysis. Breast Cancer Res Treat. 2011;127:1–14.
Kuhl CK, Mielcareck P, Klaschik S, et al. Dynamic breast mr imaging: are signal intensity time course data useful for differential diagnosis of enhancing lesions? 1. Radiology. 1999;211:101–10.
Kuhl CK, Schild HH. Dynamic image interpretation of MRI of the breast. J Magn Reson Imaging. 2000;12:965–74.
American College of Radiology (2013) Breast imaging reporting and data system (BI-RADS), 5th ed. Reston American College of Radiology
Chan S, Chen J-H, Agrawal G, et al. Characterization of pure ductal carcinoma in situ on dynamic contrast-enhanced MR Imaging: do nonhigh grade and high grade show different imaging features? J Oncol. 2010;2010:431341.
Jansen SA, Newstead GM, Abe H, et al. Pure ductal carcinoma in situ: kinetic and morphologic MR characteristics compared with mammographic appearance and nuclear grade1. Radiology. 2007;245:684–91.
Greenwood HI, Wilmes LJ, Kelil T, et al. Role of breast MRI in the evaluation and detection of DCIS: opportunities and challenges. J Magn Reson Imaging. 2020;52:697–709.
Mann RM, Mus RD, van Zelst J, et al. A novel approach to contrast-enhanced breast magnetic resonance imaging for screening: high-resolution ultrafast dynamic imaging. Invest Radiol. 2014;49:579–85.
Pineda FD, Medved M, Wang S, et al. Ultrafast bilateral DCE-MRI of the breast with conventional fourier sampling: preliminary evaluation of semi-quantitative analysis. Acad Radiol. 2016;23:1137–44.
Mori N, Abe H, Mugikura S, et al. Ultrafast dynamic CONTRAST-ENHANCED BREAST MRI: kinetic curve assessment using empirical mathematical model validated with histological microvessel density. Acad Radiol. 2019;26:e141–9.
Onishi N, Sadinski M, Gibbs P, et al. Differentiation between subcentimeter carcinomas and benign lesions using kinetic parameters derived from ultrafast dynamic contrast-enhanced breast MRI. Eur Radiol. 2020;30:756–66.
Abe H, Mori N, Tsuchiya K, et al. Kinetic analysis of benign and malignant breast lesions with ultrafast dynamic contrast-enhanced MRI: comparison with standard kinetic assessment. Am J Roentgenol. 2016;207:1159–66.
Mori N, Pineda FD, Tsuchiya K, et al. Fast temporal resolution dynamic contrast-enhanced MRI: histogram analysis versus visual analysis for differentiating benign and malignant breast lesions. Am J Roentgenol. 2018;211:933–9.
Ohashi A, Kataoka M, Kanao S, et al. Diagnostic performance of maximum slope: a kinetic parameter obtained from ultrafast dynamic contrast-enhanced magnetic resonance imaging of the breast using k-space weighted image contrast (KWIC). Eur J Radiol. 2019;118:285–92.
Facius M, Renz DM, Neubauer H, et al. Characteristics of ductal carcinoma in situ in magnetic resonance imaging. Clin Imaging. 2007;31:394–400.
Esserman LJ. Magnetic resonance imaging captures the biology of ductal carcinoma in situ. J Clin Oncol. 2006;24:4603–10.
Yoon H-J, Kim Y, Kim BS. Intratumoral metabolic heterogeneity predicts invasive components in breast ductal carcinoma in situ. Eur Radiol. 2015;25:3648–58.
Ueno Y, Forghani B, Forghani R, et al. Endometrial carcinoma: MR imaging–based texture model for preoperative risk stratification—a preliminary analysis. Radiology. 2017;284:748–57.
Yamada I, Miyasaka N, Kobayashi D, et al. Endometrial carcinoma: texture analysis of apparent diffusion coefficient maps and its correlation with histopathologic findings and prognosis. Radiol Imaging Cancer. 2019;1:e190054. https://doi.org/10.1148/rycan.2019190054.
RM Haralick, Shanmugam K, Dinstein I (1973) Textural features for image classification. IEEE Trans Syst Man Cybern (6):610–21
Gibbs P, Turnbull LW. Textural analysis of contrast-enhanced MR images of the breast. Magn Reson Med. 2003;50:92–8.
Chen W, Giger ML, Li H, et al. Volumetric texture analysis of breast lesions on contrast-enhanced magnetic resonance images. Magn Reson Med. 2007;58:562–71.
Woods BJ, Clymer BD, Kurc T, et al. Malignant-lesion segmentation using 4D co-occurrence texture analysis applied to dynamic contrast-enhanced magnetic resonance breast image data. J Magn Reson Imaging. 2007;25:495–501.
Eun NL, Kang D, Son EJ, et al. Texture analysis with 3.0-T MRI for association of response to neoadjuvant chemotherapy in breast cancer. Radiology. 2020;294:31–41.
Waugh SA, Purdie CA, Jordan LB, et al. Magnetic resonance imaging texture analysis classification of primary breast cancer. Eur Radiol. 2016;26:322–30.
Wu M, Krishna S, Thornhill RE, et al. Transition zone prostate cancer: logistic regression and machine-learning models of quantitative ADC, shape and texture features are highly accurate for diagnosis: Machine-Learning Diagnosis of PZ PCa. J Magn Reson Imaging. 2019;50:940–50.
WHO Classification of Tumours, 5th ed. Breast Tumours WORLD HEALTH ORGANIZATION, 2019
Chen W, Giger ML, Bick U. A fuzzy C-means (FCM)-based approach for computerized segmentation of breast lesions in dynamic contrast-enhanced MR Images1. Acad Radiol. 2006;13:63–72.
Alhamzawi R, Ali HTM. The Bayesian adaptive lasso regression. Math Biosci. 2018;303:75–82.
Zheng Y, Li J, Liu S, et al. MRI-Based radiomics nomogram for differentiation of benign and malignant lesions of the parotid gland. Eur Radiol. 2020. https://doi.org/10.1007/s00330-020-07483-4 ((online ahead of print)).
Yan P-F, Yan L, Hu T-T, et al. The potential value of preoperative MRI texture and shape analysis in grading meningiomas: a preliminary investigation. Transl Oncol. 2017;10:570–7.
Hamerla G, Meyer H-J, Schob S, et al. Comparison of machine learning classifiers for differentiation of grade 1 from higher gradings in meningioma: a multicenter radiomics study. Magn Reson Imaging. 2019;63:244–9.
Lehotska V, Rauova K, Vanovcanova L. Multiparametric MRI analysis of morphologico-functional features of DCIS—correlation with the grade of nuclear atypia. Neoplasma. 2018;65:389–97.
Hajaj M, Karim A, Pascaline S, et al. Impact of MRI on high grade Ductal Carcinoma Insitu (HG DCIS) management, are we using the full scope of MRI? Eur J Radiol. 2017;95:271–7.
Lee KH, Han JW, Kim EY, et al. Predictive factors for the presence of invasive components in patients diagnosed with ductal carcinoma in situ based on preoperative biopsy. BMC Cancer. 2019;19:1201.
Iima M, Bihan DL, Okumura R, et al. Apparent diffusion coefficient as an MR imaging biomarker of low-risk ductal carcinoma in situ: a pilot study. Radiology. 2011;260:364–72.
Mori N, Ota H, Mugikura S, et al. Detection of invasive components in cases of breast ductal carcinoma in situ on biopsy by using apparent diffusion coefficient MR parameters. Eur Radiol. 2013;23:2705–12.
Burstein HJ, Polyak K, Wong JS, et al. Ductal carcinoma in situ of the breast. N Engl J Med. 2004;350:1430–41.
Mannu GS, Groen EJ, Wang Z, et al. Reliability of preoperative breast biopsies showing ductal carcinoma in situ and implications for non-operative treatment: a cohort study. Breast Cancer Res Treat. 2019;178:409–18.
Mossa-Basha M, Fundaro GM, Shah BA, et al (2020) Ductal carcinoma in situ of the breast: MR imaging findings with histopathologic correlation. Radiographics 30:1673–1687.
Neubauer H. High grade and non-high grade ductal carcinoma in situ on dynamic MR mammography: characteristic findings for signal increase and morphological pattern of enhancement. Br J Radiol. 2003;76:3–12.
Rosen EL, Smith-Foley SA, DeMartini WB, et al. BI-RADS MRI enhancement characteristics of ductal carcinoma in situ. Breast J. 2007;13:545–50.
Orel SG, Mendonca M, Sullivan D. MR imaging of ductal carcinoma in situ. Radiology. 1997;202:413–20.
Acknowledgements
This research was supported by Segal foundation grant and JSPS KAKENHI (18K07742). The authors thank Robert Tomek of Qlarity Imaging for his kind support. The authors thank Naoko Hirose, Kanako Shibui, and Kyuhei Takahashi in Tohoku University for their kind support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Shape features
Volume is the volume of the lesion in mm3.
Surface area is the surface area of the lesion in mm2.
Texture features
The following mathematical expressions are used to calculate the GLCM-based texture
GLCM textures are calculated using the following equations:
where \({\mu }_{x-y}\) is the mean of \({p}_{x-y}(k)\)
where
where N* is the number of voxels in the border, B*, and Δxyz is the isotropic voxel size.
Where
were
About this article
Cite this article
Mori, N., Abe, H., Mugikura, S. et al. Discriminating low-grade ductal carcinoma in situ (DCIS) from non-low-grade DCIS or DCIS upgraded to invasive carcinoma: effective texture features on ultrafast dynamic contrast-enhanced magnetic resonance imaging. Breast Cancer 28, 1141–1153 (2021). https://doi.org/10.1007/s12282-021-01257-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-021-01257-6