Abstract
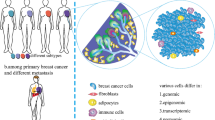
Intricacy in treatment and diagnosis of breast cancer has been an obstacle due to genotype and phenotype heterogeneity. Understanding of non-genetic heterogeneity mechanisms along with considering role of genetic heterogeneity may fill the gaps in landscape painting of heterogeneity. The main factors contribute to non-genetic heterogeneity including: transcriptional pulsing/bursting or discontinuous transcriptions, stochastic partitioning of components at cell division and various signal transduction from tumor ecosystem. Throughout this review, we desired to provide a conceptual framework focused on non-genetic heterogeneity, which has been intended to offer insight into prediction, diagnosis and treatment of breast cancer.

source of the heterogeneity. Transcriptional pulsing/bursting or discontinuous transcriptions, stochastic partitioning of components at cell division belong to intrinsic sources of non-genetic phenotypic and functional heterogeneity. Various signal transduction from TME in line with variation in TME component and topography cause extrinsic source of non-genetic heterogeneity in cancer



Similar content being viewed by others
Abbreviations
- CAFs:
-
Cancer-associated fibroblasts
- TADs:
-
Topologically associating domains
- CTCF:
-
CCCTC-binding factor
- lncRNAs:
-
Long noncoding RNA
- DMGs:
-
Methylated DNA regions
- MDSCs:
-
Myeloid-derived suppressor cells
- TAMs:
-
Tumor-associated macrophages
- VECs:
-
Vascular endothelial cells
- Program death ligand 1:
-
Expressing PD-L1
- EMT/MET:
-
Epithelial to mesenchymal/mesenchymal to epithelial transition
- CSCs:
-
Cancer stem-like cells
- TME:
-
Tumor microenvironment
- ICD:
-
Immunogenic cell death
- HMGB1:
-
High-mobility group box 1 protein
- HIF1α:
-
Hypoxia inducible factor 1 alpha
- CTCs:
-
Circulating tumor cells
- ctDNA:
-
Circulating tumor DNA
- EVs:
-
Extracellular vesicles
- MCT1:
-
Monocarboxylate transporter 1
- BCSCs:
-
Breast cancer CSCs
- CK14:
-
Cytokeratin-14
- PDGF:
-
Platelet-derived growth factor
- NSCCs:
-
Non-stem cancer cells
- IHC:
-
Immunohistochemistry
- IF:
-
Immunofluorescence
- MERFISH:
-
Multiplexed error robust FISH
- WGA:
-
Genome analysis
- NGS:
-
Next-generation sequence
- FACS:
-
Fluorescence-activated cell sorting
- WTS:
-
Whole transcriptome sequencing
- CyTOF:
-
Cytometry time of flight
- MALDI:
-
Matrix-assisted laser desorption/ionization
References
Lee ATJ, Chew W, Wilding CP, Guljar N, Smith MJ, Strauss DC, et al. The adequacy of tissue microarrays in the assessment of inter- and intra-tumoural heterogeneity of infiltrating lymphocyte burden in leiomyosarcoma. Sci Rep. 2019;9:1–12.
Cornwell JA, Hallett RM, Der MSA, Motazedian A, Schroeder T. Quantifying intrinsic and extrinsic control of single-cell fates in cancer and stem/progenitor cell pedigrees with competing risks analysis. Sci Rep. 2016. https://doi.org/10.1038/srep27100.
Kærn M, Elston TC, Blake WJ, Collins JJ. Stochasticity in gene expression: from theories to phenotypes. Nat Rev Genet. 2005;6:451–64.
Thomas P. Intrinsic and extrinsic noise of gene expression in lineage trees. Sci Rep. 2019. https://doi.org/10.1038/s41598-018-35927-x.
Kwon T, Kwon O, Cha H, Sung BJ. Stochastic and heterogeneous cancer cell migration: experiment and theory. Sci Rep. 2019;9:1–13.
Voss TC, Hager GL. Dynamic regulation of transcriptional states by chromatin and transcription factors. Nat Rev Genet. 2013. https://doi.org/10.1038/nrg3623.
Lahav G, Rosenfeld N, Sigal A, Geva-zatorsky N, Levine AJ, Elowitz MB, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nature. 2004;36(2):147–50.
Sh C, Lo A. p53 dynamics in single cells are. 2020;1–14.
Chubb JR. Gene regulation: stable noise. Curr Biol. 2016;26(2):R61–4. https://doi.org/10.1016/j.cub.2015.12.002.
Huang S. Non-genetic heterogeneity of cells in development: more than just noise. Development. 2009;3862:3853–62.
Huh D, Paulsson J. Non-genetic heterogeneity from stochastic partitioning at cell division. Nat Genet. 2011;43(2):95.
Mulder N, Martin DP. Metabolic gene alterations impact the clinical aggressiveness and drug responses of 32 human cancers. Commun Biol. 2019. https://doi.org/10.1038/s42003-019-0666-1.
Tonn MK, Thomas P, Oyarzún DA. Stochastic modelling reveals mechanisms of metabolic heterogeneity. Commun Biol. 2019. https://doi.org/10.1038/s42003-019-0347-0.
Wilkinson M, Darmanis S, Pisco AO, Huber G. Persistent features of intermittent transcription. Sci Rep. 2020. https://doi.org/10.1038/s41598-020-60094-3.
Sahai E, Astsaturov I, Cukierman E, Denardo DG, Egeblad M, Evans RM, et al. A framework for advancing our understanding of cancer-associated fibroblasts. Nat Rev Cancer. 2020. https://doi.org/10.1038/s41568-019-0238-1.
Krivega I, Dean A. CTCF fences make good neighbours. Nat Cell Biol. 2017;19(8):883–5. https://doi.org/10.1038/ncb3584.
Ghavi-Helm Y, Jankowski A, Meiers S, Viales RR, Korbel JO, Furlong EEM. Highly rearranged chromosomes reveal uncoupling between genome topology and gene expression. Nat Genet. 2019. https://doi.org/10.1038/s41588-019-0462-3.
Li L, Lyu X, Qin ZS, Corces VG. Widespread rearrangement of 3D chromatin organization underlies polycomb-mediated stress-induced silencing. Mol Cell. 2015;28:1–16.
Gong Y, Lazaris C, Sakellaropoulos T, Lozano A, Kambadur P, Ntziachristos P, et al. Stratification of TAD boundaries reveals preferential insulation of super-enhancers by strong boundaries. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-03017-1.
Bell CC. Principles and mechanisms of non-genetic resistance in cancer. Br J Cancer. 2019. https://doi.org/10.1038/s41416-019-0648-6.
Gelato KA, Shaikhibrahim Z, Ocker M, Haendler B. Targeting epigenetic regulators for cancer therapy: modulation of bromodomain proteins, methyltransferases, demethylases, and microRNAs. Expert Opin Ther Targets. 2016;20:783.
Wong EM, Southey MC, Terry MB. Integrating DNA methylation measures to improve clinical risk assessment: are we there yet ? The case of BRCA1 methylation marks to improve clinical risk assessment of breast cancer. Br J Cancer. 2020. https://doi.org/10.1038/s41416-019-0720-2.
Hansen KD, Timp W, Bravo HC, Sabunciyan S, Langmead B, Mcdonald OG, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43(8):768–75. https://doi.org/10.1038/ng.865.
San R, Sally AM. Integrative network analysis of differentially methylated and expressed genes for biomarker identification in leukemia. Sci Rep. 2020;10:1–16.
Moufarrij S, Srivastava A, Gomez S, Hadley M, Palmer E, Austin PT, et al. Combining DNMT and HDAC6 inhibitors increases anti-tumor immune signaling and decreases tumor burden in ovarian cancer. Sci Rep. 2020;10:1–12.
Barrett RDH. Epigenetics in natural animal populations. J Evol Biol. 2017;30:1612–32.
Burkhart RA, Laheru DA, Herman JM, Timothy M. Multidisciplinary management and the future of treatment in cholangiocarcinoma. Expert Opin Orphan Drugs. 2016;4:255.
Wang G, Wang Q, Liang N, Xue H, Yang T, Chen X, et al. Oncogenic driver genes and tumor microenvironment determine the type of liver cancer. Cell Death Dis. 2020. https://doi.org/10.1038/s41419-020-2509-x.
Po A, Giuliani A, Masiello MG, Cucina A, Chiacchiarini M, Tafani M, et al. Phenotypic transitions enacted by simulated microgravity do not alter coherence in gene transcription profile. npj Microgravity. 2019. https://doi.org/10.1038/s41526-019-0088-x.
Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74.
Liu S, Qin T, Liu Z, Wang J, Jia Y, Feng Y, et al. anlotinib alters tumor immune microenvironment by downregulating PD-L1 expression on vascular endothelial cells. Cell Death Dis. 2020. https://doi.org/10.1038/s41419-020-2511-3.
Thibaut R, Bost P, Milo I, Cazaux M, Lemaître F, Garcia Z, et al. Bystander IFN-γ activity promotes widespread and sustained cytokine signaling altering the tumor microenvironment. Nat Cancer. 2020. https://doi.org/10.1038/s43018-020-0038-2.
Kim MH, Kim J, Lee JM, Choi JW, Jung D, Cho H, et al. Molecular subtypes of oropharyngeal cancer show distinct immune microenvironment related with immune checkpoint blockade response. Br J Cancer. 2020. https://doi.org/10.1038/s41416-020-0796-8.
Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, et al. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Targeted Ther. 2020. https://doi.org/10.1038/s41392-020-0110-5.
Vickers NJ. Animal communication : When I’m Calling You, Will You Answer Too? Curr Biol. 2017;27(14):R713–5. https://doi.org/10.1016/j.cub.2017.05.064.
Wilcken N, Zdenkowski N, White M, Snyder R, Pittman K, Mainwaring P, et al. Systemic treatment of HER2-positive metastatic breast cancer: a systematic review. Asia Pac J Clin Oncol. 2014;10:1–14.
Ohlsson R, Kanduri C, Whitehead J, Pfeifer S, Lobanenkov V, Feinberg AP. Epigenetic variability and the evolution of human cancer. Adv Cancer Res. 2003;88:145.
Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33.
Feinberg AP, Koldobskiy MA, Göndör A. Disease mechanisms: epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet. 2016. https://doi.org/10.1038/nrg.2016.13.
Zhang Y. Epithelial-to-mesenchymal transition in cancer: complexity and opportunities. Front Med. 2018;12(4):361–73.
Papadaki MA, Stoupis G, Theodoropoulos PA, Mavroudis D. Circulating tumor cells with stemness and epithelial-to-mesenchymal transition features are chemoresistant and predictive of poor outcome in metastatic breast cancer. Mol Cancer. 2019;18:437–48.
Bhang HC, Ruddy DA, Radhakrishna VK, Caushi JX, Zhao R, Hims MM, et al. Articles studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat Med. 2015;21(5):440.
Frankenstein Z, Basanta D, Franco OE, Gao Y, Javier RA, Strand DW, et al. Stromal reactivity differentially drives tumour cell evolution and prostate cancer progression. Nat Ecol Evol. 2020. https://doi.org/10.1038/s41559-020-1157-y.
Junttila MR, De SFJ. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 2013;501:346.
Kalluri R. The biology and function of fibroblasts in cancer. Nat Rev. 2016;16(9):582–98. https://doi.org/10.1038/nrc.2016.73.
Cairns J, Ung CY, Lummertz E, Zhang C, Correia C, Weinshilboum R, et al. A network-based phenotype mapping approach to identify genes that modulate drug response phenotypes. Sci Rep. 2016;6:1–13.
Sun Y, Campisi J, Higano C, Beer TM, Porter P, Coleman I, et al. Treatment-induced damage to the tumor micro-environment promotes prostate cancer therapy resistance through WNT16B. Nat Med. 2012;18(9):1359–68. https://doi.org/10.1038/nm.2890.
Takeshima H, Ushijima T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. npj Precis Oncol. 2019. https://doi.org/10.1038/s41698-019-0079-0.
Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1 β—dependent adaptive immunity against tumors. Nat Med. 2009;15(10):1170–9.
Kroemer G, Galluzzi L, Kepp O, Zitvogel L. Immunogenic cell death in cancer therapy. Annu Rev Ther. 2013;31:51.
Ye Y, Hu Q, Chen H, Liang K, Yuan Y, Xiang Y, et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat Metab. 2019. https://doi.org/10.1038/s42255-019-0045-8.
Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet. 2019. https://doi.org/10.1038/s41588-018-0318-2.
Dai X, Xiang L, Li T, Bai Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J Cancer. 2016;7(10):1281–94.
Geng H, Xue C, Mendonca J, Sun X, Liu Q, Reardon PN, et al. Interplay between hypoxia and androgen controls a metabolic switch conferring resistance to androgen/AR-targeted therapy. Nat Commun. 2018. https://doi.org/10.1038/s41467-018-07411-7.
Raja R, Kale S, Thorat D, Soundararajan G, Lohite K, Mane A, et al. Hypoxia-driven osteopontin contributes to breast tumor growth through modulation of HIF1 a-mediated VEGF-dependent angiogenesis. Oncogene. 2014;33:2053–64.
Kim H, Lin Q, Yun Z. BRCA1 regulates the cancer stem cell fate of breast cancer cells in the context of hypoxia and histone deacetylase inhibitors. Sci Rep. 2019. https://doi.org/10.1038/s41598-019-46210-y.
Barrak NH, Khajah MA, Luqmani YA. Hypoxic environment may enhance migration/penetration of endocrine resistant MCF7-derived breast cancer cells through monolayers of other non-invasive cancer cells in vitro. Sci Rep. 2020. https://doi.org/10.1038/s41598-020-58055-x.
Inference B, Farlik M, Sheffield NC, Klughammer J, Bock C, Klughammer J. Single-cell DNA methylome sequencing and resource single-cell DNA methylome sequencing and bioinformatic inference of epigenomic cell-state dynamics. Cell Rep. 2015;10:1386–97.
Conley SJ, Gheordunescu E, Kakarala P, Newman B, Korkaya H, Heath AN, et al. Antiangiogenic agents increase breast cancer stem cells via the generation of tumor hypoxia. Proc Natl Acad Sci. 2012;109(8):2784.
Widmer DS, Hoek KS, Cheng PF, Eichhoff OM, Biedermann T, Raaijmakers MIG, et al. Hypoxia contributes to melanoma heterogeneity by triggering HIF1 a-dependent phenotype switching. J Investig Dermatol. 2013;133:2436.
Thalgott M, Rack B, Maurer T, Souvatzoglou M, Eiber M, Kreß V, et al. Detection of circulating tumor cells in different stages of prostate cancer. J Cancer Res Clin Oncol. 2013;139:755–63.
Yu M, Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, et al. 2013;580.
Follain G, Herrmann D, Hyenne V, Warren SC, Timpson P, Goetz JG. Fluids and their mechanics in tumour transit: shaping metastasis. Nat Rev. 2020;20:107.
Headley MB, Bins A, Nip A, Edward W, Looney MR, Gerard A, et al. Visualization of immediate immune responses to pioneer metastatic cells in the lung. Nature. 2016. https://doi.org/10.1038/nature16985.
Azevedo AS, Pantel K, Goetz JG, Metivet T, Hille C, Chabannes V, et al. Hemodynamic forces tune the arrest, adhesion, and extravasation of circulating tumor cells. Dev Cells. 2018;45:33–52.
Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, et al. Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature. 2019. https://doi.org/10.1038/s41586-019-1847-2.
Gkountela S, Castro-giner F, Szczerba BM, Rochlitz C, Weber WP, Gkountela S, et al. Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding article circulating tumor cell clustering shapes DNA methylation. Cell. 2019;176(1–2):98–112. https://doi.org/10.1016/j.cell.2018.11.046.
Ignatiadis M, Lee M, Jeffrey SS. Circulating tumor cells and circulating tumor DNA: challenges and opportunities on the path to clinical utility. Clin Cancer Res. 2015;21(21):4786–801.
Tricarico C, Clancy J, Souza-schorey CD. Biology and biogenesis of shed microvesicles. Small GTPases. 2017;8(4):220–32. https://doi.org/10.1080/21541248.2016.1215283.
Keklikoglou I, Cianciaruso C, Güç E, Squadrito ML, Spring LM, Tazzyman S, et al. Chemotherapy elicits pro-metastatic extracellular vesicles in breast cancer models. Nat Cell Biol. 2019;21(2):190–202.
Bandura DR, Baranov VI, Ornatsky OI, Antonov A, Kinach R, Lou X, et al. Mass cytometry: technique for real time single cell multitarget immunoassay based on inductively coupled plasma time-of-flight mass spectrometry. Anal Chem. 2016;81(16):6813–22.
Eun K, Ham SW, Kim H. Cancer stem cell heterogeneity: origin and new perspectives on CSC targeting. BMB Rep. 2017;50(3):117–25.
Chen W, Dong J, Haiech J, Kilhoffer M, Zeniou M. Cancer stem cell quiescence and plasticity as major challenges in cancer therapy. Stem Cells Int. 2016.
Santisteban M, Reiman JM, Asiedu MK, Behrens MD, Nassar A, Kalli KR, et al. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res. 2009;69(7):2887–96.
Sharma A, Merritt E, Hu X, Malhotra J, Riedlinger GM, De S, et al. Non-genetic intra-tumor heterogeneity is a major predictor of phenotypic heterogeneity and ongoing evolutionary dynamics in lung tumors. Cell Rep. 2019;29(8):2164–74. https://doi.org/10.1016/j.celrep.2019.10.045.
He P, Qiu K, Jia Y. Modeling of mesenchymal hybrid epithelial state and phenotypic transitions in EMT and MET processes of cancer cells. Sci Rep. 2018;8:1–13.
Mani SA, Guo W, Liao M, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial–mesenchymal transition generates cells with properties of stem cells. Cells. 2008;133:704–15.
Sun L, Burnett J, Gasparyan M, Xu F, Jiang H, Lin C, et al. Novel cancer stem cell targets during epithelial to mesenchymal transition in PTEN-deficient trastuzumab-resistant breast cancer. Oncotarget. 2016;7(32):51408.
Almendro V, Marusyk A, Polyak K. Cellular heterogeneity and molecular evolution in cancer. Annu Rev Pathol Mech Dis. 2013;8:277.
Qiu K, Gao K, Yang L, Zhang Z, Wang R. OPEN A kinetic model of multiple phenotypic states for breast cancer cells. Sci Rep. 2017. https://doi.org/10.1038/s41598-017-10321-1.
Gupta PB, Fillmore CM, Jiang G, Shapira SD, Tao K, Kuperwasser C. Theory stochastic state transitions give rise to phenotypic equilibrium in populations of cancer cells. Cell. 2011;146(4):633–44. https://doi.org/10.1016/j.cell.2011.07.026.
Yang C, Cao M, Liu Y, He Y, Du Y, Zhang G, et al. Inducible formation of leader cells driven by CD44 switching gives rise to collective invasion and metastases in luminal breast carcinomas. Oncogene. 2019. https://doi.org/10.1038/s41388-019-0899-y.
Cheung KJ, Gabrielson E, Werb Z, Ewald AJ. Collective invasion in breast cancer requires a conserved basal epithelial program. Cell. 2013;155(7):1639–51. https://doi.org/10.1016/j.cell.2013.11.029.
Jordan NV, Bardia A, Wittner BS, Benes C, Ligorio M, Zheng Y, et al. HER2 expression identifies dynamic functional states within circulating breast cancer cells. Nature. 2016. https://doi.org/10.1038/nature19328.
Roswall P, Bocci M, Bartoschek M, Li H, Kristiansen G, Jansson S, et al. Microenvironmental control of breast cancer subtype elicited through paracrine platelet-derived growth factor-CC signaling. Nat Med. 2018. https://doi.org/10.1038/nm.4494.
Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci. 2011;108(4):1397–402.
Aparicio S, Caldas C. The implications of clonal genome evolution for cancer medicine. N Engl J Med. 2013;368:842.
Bassez A, Decaluwé H, Pircher A, Van Den EK. Tumor microenvironment. Nat Med. 2018. https://doi.org/10.1038/s41591-018-0096-5.
Learned L, Challenges E. Perspective single-cell RNA sequencing in cancer: lessons learned and emerging challenges. Mol Cell. 2019;75:7–12.
Martelotto LG, Ng CKY, Piscuoglio S, Weigelt B, Reis-filho JS. Breast cancer intra-tumor heterogeneity. Breast Cancer Res. 2014;16:1.
Itzkovitz S, Van OA. Validating transcripts with probes and imaging technology. Nat Methods. 2011;8(4):S12.
Massard C, Oulhen M, Le MS, Foulon S, Abou-lovergne A, Billiot F. Phenotypic and genetic heterogeneity of tumor tissue and circulating tumor cells in patients with metastatic castration-resistant prostate cancer: a report from the PETRUS prospective study. Oncotarget. 2016;7(34):55069.
Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, Wu X, Vo HT, Ma XJ, Luo Y. RNAscope: a novel in situ RNA analysis platform for formalin fixed, paraffin-embedded tissues. J Mol Diagn. 2012;14(1):22–9.
Park SY, Michor F, Polyak K, Park SY, Gönen M, Kim HJ, et al. Cellular and genetic diversity in the progression of in situ human breast carcinomas to an invasive phenotype. J Clin Investig. 2010;120(2):636–44.
Chen KH, Boettiger AN, Moffitt JR, Wang S, Zhuang X. Spatially resolved, highly multiplexed RNA profiling in single cells. Science. 2015;1363(2014):1360–3.
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, Mcindoo J, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472(7341):90–4. https://doi.org/10.1038/nature09807.
Powell AA, Talasaz AH, Zhang H, Coram MA, Reddy A, Deng G, Telli ML, Advani RH, Carlson RW, Mollick JA, Sheth S. Single cell profiling of circulating tumor cells: transcriptional heterogeneity and diversity from breast cancer cell lines. PloS One. 2012;7(5):e33788.
Sidoli S, Kori Y, Lopes M, Yuan Z, Kim HJ, Kulej K, et al. One minute analysis of 200 histone posttranslational modifications by direct injection mass spectrometry. Genome Res. 2019;29:978–87.
Dedeurwaerder S, Desmedt C, Calonne E, Singhal SK, Haibe-kains B, Defrance M, et al. DNA methylation profiling reveals a predominant immune component in breast cancers. EMBO Mol Med. 2011;3:726–41.
Cancer T, Atlas G. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:1–10.
Lang JE, Scott JH, Wolf DM, Novak P, Punj V, Jesus M, et al. Expression profiling of circulating tumor cells in metastatic breast cancer. Breast Cancer Res Treat. 2014;149:121.
Doherty R, Couldrey C, Huang W, State NC. Exploring genome wide bisulfite sequencing for DNA methylation analysis in livestock: a technical assessment. Front Genet. 2014;5:1–8.
Wen L, Tang F. Single-cell sequencing in stem cell biology. Genome Biol. 2016. https://doi.org/10.1186/s13059-016-0941-0.
Saliba A, Westermann AJ, Gorski SA. Single-cell RNA-seq: advances and future challenges. Nucleic Acids Res. 2014;42(14):8845–60.
Wlodkowic D, Darzynkiewicz Z. Rise of the micromachines: microfluidics and the future of cytometry. Methods Cell Biol. 2011;102:105–25. https://doi.org/10.1016/B978-0-12-374912-3.00005-5.
Irish JM, Myklebust JH, Alizadeh AA, Houot R, Sharman JP, Czerwinski DK. B-Cell signaling networks reveal a negative prognostic human lymphoma cell subset that emerges during tumor progression. Proc Natl Acad Sci. 2010;107(29):12747.
Blake LA, Wu B. One message, many translations: heterogeneity revealed with multicolor imaging. Mol cell. 2019;75(1):3–4.
Blake LA, Wu B. Previews one message, many translations: heterogeneity revealed with multicolor imaging. Mol Cell. 2019;75(1):3–4. https://doi.org/10.1016/j.molcel.2019.06.026.
Alizadeh AA, Aranda V, Bardelli A, Blanpain C, Bock C, Borowski C, et al. Toward understanding and exploiting tumor heterogeneity. Nat Med. 2015;21(8):1–8. https://doi.org/10.1038/nm.3915.
Mayor S. Swiss vote “no” to comprehensive smoking ban. Lancet Oncol. 2012;13(11):e466. https://doi.org/10.1016/S1470-2045(12)70439-4.
Leary RJ, Kinde I, Diehl F, Schmidt K, Clouser C, Duncan C, et al. Development of personalized tumor biomarkers using massively parallel sequencing. Sci Transl Med. 2010;2(20):1–8.
Seol H, Lee HJ, Choi Y, Lee HE, Kim YJ, Kim JH, et al. Intratumoral heterogeneity of HER2 gene amplification in breast cancer: its clinicopathological significance. Mod Pathol. 2012;25:938–48.
Sood A, Miller AM, Brogi E, Sui Y, Mcdonough E, Santamaria-pang A, et al. Multiplexed immunofluorescence delineates proteomic cancer cell states associated with metabolism. JCI Insight. 2016;1(6):1–14.
Meyer AS, Heiser LM. Systems biology approaches to measure and model phenotypic heterogeneity in cancer. Curr Opin Syst Biol. 2019;17:35–40. https://doi.org/10.1016/j.coisb.2019.09.002.
Wen Y, Wei Y, Zhang S, Li S, Liu H, Wang F. Cell subpopulation deconvolution reveals breast cancer heterogeneity based on DNA methylation signature. Brief Bioinform. 2017;18:426–40.
Miura S, Vu T, Deng J, Buturla T, Oladeinde O. Power and pitfalls of computational methods for inferring clone phylogenies and mutation orders from bulk sequencing data. Sci Rep. 2020. https://doi.org/10.1038/s41598-020-59006-2.
Malikic S, Jahn K, Kuipers J, Sahinalp SC, Beerenwinkel N. Integrative inference of subclonal tumour evolution from single-cell and bulk sequencing da. Nat Commun. 2019. https://doi.org/10.1038/s41467-019-10737-5.
Wang N, Zheng J, Chen Z, Liu Y, Dura B, Kwak M, et al. Single-cell microRNA-mRNA co-sequencing reveals non-genetic heterogeneity and mechanisms of microRNA regulation. Nat Commun. 2019. https://doi.org/10.1038/s41467-018-07981-6.
Almendro V, Fuster G. Heterogeneity of breast cancer: etiology and clinical relevance. Clin Trans Oncol. 2011;13:767–73.
Jesneck JL, Nolte LW, Baker JA, Floyd CE, Lo JY, Nolte LW. Optimized approach to decision fusion of heterogeneous data for breast cancer diagnosis. Med Phys. 2006;33:2945.
Clark BZ, Onisko A, Assylbekova B, Li X, Bhargava R, Dabbs DJ, et al. Breast cancer global tumor biomarkers: a quality assurance study of intratumoral heterogeneity. Mod Pathol. 2018. https://doi.org/10.1038/s41379-018-0153-0.
Guiu S, Michiels S, André F, Cortes J, Denkert C, Di Leo A, et al. Molecular subclasses of breast cancer: How do we define them? The IMPAKT 2012 working group statement. Ann Oncol. 2012;23:2997–3006.
Zardavas D, Irrthum A, Swanton C, Piccart M. Clinical management of breast cancer heterogeneity. Nat Rev Clin. 2015;12(7):381–94. https://doi.org/10.1038/nrclinonc.2015.73.
Hon JDC, Singh B, Sahin A, Du G, Wang J, Wang VY. Breast cancer molecular subtypes: from TNBC to QNBC. Am J Cancer Res. 2016;6(9):1864–72.
C Presentation. Breast cancer treatment: a review. JAMA. 2019;321(3):288.
Kim J, De Sampaio PC, Lundy DM, Peng Q, Evans KW, Sugimoto H, et al. Heterogeneous perivascular cell coverage affects breast cancer metastasis and response to chemotherapy. JCI Insights. 2016;1(21):1–17.
Cai S, Allam M, Coskun AF. Forum multiplex spatial bioimaging for combination therapy design. Trends Cancer. 2020. https://doi.org/10.1016/j.trecan.2020.05.003.
Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34:378–90.
Bergenfelz C, Larsson A, Von Stedingk K, Gruvberger-Saal S, Aaltonen K, Jansson S, Jernström H, Janols H, Wullt M, Bredberg A, Rydén L. Systemic monocytic-MDSCs are generated from monocytes and correlate with disease progression in breast cancer patients. Plos One. 2015;10(5):e0127028.
Beca F, Polyak K. Intratumor heterogeneity in breast cancer. In: Stearns V, editor. Novel biomarkers in the continuum of breast cancer. Cham: Springer; 2016. p. 169–89.
Acknowledgements
This research project was financially supported by Immunology Research Center at Tabriz University of Medical Sciences. This grant supported MSC thesis of Neda Barzgar Barough (The thesis code: 97/12/14 and ethical code: IR.TBZMED.VCR.REC.1398.288).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
N.B.B., F.S., and N.J. have participated in writing of main body of review article. In addition, H. Sh. has reviewed and edited the manuscript. Finally, K.V. was responsible for conceptualization, validation of different sections and illustration of figures.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Barzgar Barough, N., Sajjadian, F., Jalilzadeh, N. et al. Understanding breast cancer heterogeneity through non-genetic heterogeneity. Breast Cancer 28, 777–791 (2021). https://doi.org/10.1007/s12282-021-01237-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-021-01237-w