Abstract
Background
Long non-coding RNA (LncRNA) Breast cancer anti-estrogen resistance 4 (BCAR4) has been shown to participate in the biological progress of various cancers including breast cancer. Genetic Polymorphisms in BCAR4 may have an influence on the progress of breast cancer, but it is rarely studied.
Methods
In our epidemiology study, three tagging SNPs (rs4561483, rs11649623 and rs13334967) in lncRNA BCAR4 were selected for genotyping among 487 breast cancer cases and 489 healthy controls. And quantitative real-time PCR (qRT-PCR) was performed to evaluate the relative mRNA expression of BCAR4 in different genotypes of the significant locus rs13334967.
Results
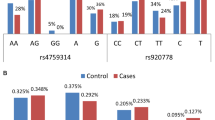
We found that BCAR4 rs13334967 is associated with lower breast cancer risk both in codominant model (AT vs AA, OR 0.632, 95% CI 0.429–0.931, TT vs AA, OR 0.731, 95% CI 0.511–0.990) and dominant model (AT + TT vs AA, OR 0.798, 95% CI 0.571–0.970). The further results of qRT-PCR displayed that carriers with rs13334967 AT, TT genotype have lower BCAR4 mRNA expression compared with AA genotype.
Conclusion
The research study implied that BCAR4 rs13334967 was correlated with the susceptibility to breast cancer and may impact the mRNA expression of BCAR4. LncRNA BCAR4 may be a potential biomarker and therapeutic target for breast cancer.

Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Fan L, Strasser-Weippl K, Li JJ, St LJ, Finkelstein DM, Yu KD, et al. Breast cancer in China. Lancet Oncol. 2014;15:e279–89.
Goss PE, Strasser-Weippl K, Lee-Bychkovsky BL, Fan L, Li J, Chavarri-Guerra Y, et al. Challenges to effective cancer control in China, India, and Russia. Lancet Oncol. 2014;15:489–538.
Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77:3965–81.
Chen LL. Linking long noncoding RNA localization and function. Trends Biochem Sci. 2016;41:761–72.
Gibb EA, Brown CJ, Lam WL. The functional role of long non-coding RNA in human carcinomas. Mol Cancer. 2011;10:38.
Hrdlickova B, de Almeida RC, Borek Z, Withoff S. Genetic variation in the non-coding genome: involvement of micro-RNAs and long non-coding RNAs in disease. Biochim Biophys Acta. 2014;1842:1910–22.
Ghafouri-Fard S, Taheri M. Colon Cancer-Associated Transcripts 1 and 2: roles and functions in human cancers. J Cell Physiol. 2019;234:14581–600.
Al-Rugeebah A, Alanazi M, Parine NR. MEG3: an oncogenic long non-coding RNA in different cancers. Pathol Oncol Res. 2019;25:859–74.
Jiang Y, Du F, Chen F, Qin N, Jiang Z, Zhou J, et al. Potentially functional variants in lncRNAs are associated with breast cancer risk in a Chinese population. Mol Carcinog. 2017;56:2048–57.
Shen C, Yan T, Wang Z, Su HC, Zhu X, Tian X, et al. Variant of SNP rs1317082 at CCSlnc362 (RP11–362K14.5) creates a binding site for miR-4658 and diminishes the susceptibility to CRC. Cell Death Dis. 2018;9:1177.
Collins FS, Brooks LD, Chakravarti A. A DNA polymorphism discovery resource for research on human genetic variation. Genome Res. 1998;8:1229–31.
Hu P, Qiao O, Wang J, Li J, Jin H, Li Z, et al. rs1859168 A > C polymorphism regulates HOTTIP expression and reduces risk of pancreatic cancer in a Chinese population. World J Surg Oncol. 2017;15:155.
Li H, Tang XM, Liu Y, Li W, Chen Q, Pan Y. Association of functional genetic variants of HOTAIR with hepatocellular carcinoma (HCC) susceptibility in a Chinese population. Cell Physiol Biochem. 2017;44:447–54.
Meijer D, van Agthoven T, Bosma PT, Nooter K, Dorssers LC. Functional screen for genes responsible for tamoxifen resistance in human breast cancer cells. Mol Cancer Res. 2006;4:379–86.
Xing Z, Lin A, Li C, Liang K, Wang S, Liu Y, et al. lncRNA directs cooperative epigenetic regulation downstream of chemokine signals. Cell. 2014;159:1110–25.
Zhao W, Wang Z, Fang X, Li N, Fang J. Long noncoding RNA Breast cancer antiestrogen resistance 4 is associated with cancer progression and its significant prognostic value. J Cell Physiol. 2018;8:12956–63.
Yang H, Yan L, Sun K, Sun X, Zhang X, Cai K, et al. lncRNA BCAR4 increases viability, invasion, and migration of non-small cell lung cancer cells by targeting glioma-associated oncogene 2 (GLI2). Oncol Res. 2019;27:359–69.
Shui X, Zhou C, Lin W, Yu Y, Feng Y, Kong J. Long non-coding RNA BCAR4 promotes chondrosarcoma cell proliferation and migration through activation of mTOR signaling pathway. Exp Biol Med (Maywood). 2017;242:1044–50.
Li N, Gao WJ, Liu NS. LncRNA BCAR4 promotes proliferation, invasion and metastasis of non-small cell lung cancer cells by affecting epithelial-mesenchymal transition. Eur Rev Med Pharmacol Sci. 2017;21:2075–86.
Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. 2016;20:4445–51.
Wang L, Chunyan Q, Zhou Y, He Q, Ma Y, Ga Y, et al. BCAR4 increase cisplatin resistance and predicted poor survival in gastric cancer patients. Eur Rev Med Pharmacol Sci. 2017;21:4064–70.
Godinho M, Meijer D, Setyono-Han B, Dorssers LC, van Agthoven T. Characterization of BCAR4, a novel oncogene causing endocrine resistance in human breast cancer cells. J Cell Physiol. 2011;226:1741–9.
Xing Z, Park PK, Lin C, Yang L. LncRNA BCAR4 wires up signaling transduction in breast cancer. RNA Biol. 2015;12:681–9.
Zheng X, Han H, Liu GP, Ma YX, Pan RL, Sang LJ, et al. LncRNA wires up Hippo and Hedgehog signaling to reprogramme glucose metabolism. EMBO J. 2017;36:3325–35.
Godinho MF, Wulfkuhle JD, Look MP, Sieuwerts AM, Sleijfer S, Foekens JA, et al. BCAR4 induces antioestrogen resistance but sensitises breast cancer to lapatinib. Br J Cancer. 2012;107:947–55.
Minotti L, Agnoletto C, Baldassari F, Corra F, Volinia S. SNPs and somatic mutation on long non-coding RNA: new frontier in the cancer studies? High Throughput. 2018;7:34.
Sabarinathan R, Tafer H, Seemann SE, Hofacker IL, Stadler PF, Gorodkin J. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013;41:W475–9.
Wapinski O, Chang HY. Long noncoding RNAs and human disease. Trends Cell Biol. 2011;21:354–61.
Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–63.
Fletcher O, Dudbridge F. Candidate gene-environment interactions in breast cancer. BMC Med. 2014;12:195.
Acknowledgements
The authors would like to thank the participants, the coordinators, and administrators for their supports during the study.
Funding
This work was supported by Henan university science and technology innovation talents support program (19HASTIT005); Medical Science and Technology key projects of Henan Province and Zhengzhou (192102310088, 19A32000820, SBGJ2018089). The National Natural Science Foundation of China (U1604168).
Author information
Authors and Affiliations
Contributions
Conceptualization: RP, JC, QG, QS, LX, XX, CS. Validation: RP, JC, XX, CS. Formal analysis: RP, JC. Investigation: RP, JC. Writing-original draft: RP, JC. Writing-review and editing: RP, JC, QG, QS, LX, XX, CS. Visualization: QG, QS, LX. Supervision: XX, CS. Project administration: XX, CS.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Ethical responsibilities of authors
The manuscript has not been submitted to more than one journal for simultaneous consideration. The manuscript has not been published previously (partly or in full). A single study is not split up into several parts to increase the quantity of submissions and submitted to various journals or to one journal over time. No data have been fabricated or manipulated (including images) to support your conclusions. No data, text, or theories by others are presented as if they were the author’s own.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Peng, R., Cao, J., Guo, Q. et al. Variant in BCAR4 gene correlated with the breast cancer susceptibility and mRNA expression of lncRNA BCAR4 in Chinese Han population. Breast Cancer 28, 424–433 (2021). https://doi.org/10.1007/s12282-020-01174-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-020-01174-0