Abstract
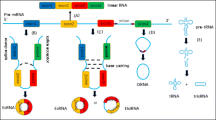
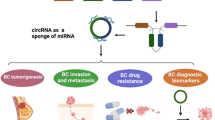
Circular RNAs are a large group of non-coding RNAs with a closed-loop structure. circRNAs play significant roles in many biological processes as miRNA sponges, regulators for gene transcription, combining with RNA-binding proteins and translation of protein. Nowadays, circRNAs have become a research hotspot in the field of cancer and molecular biology. Accumulating evidences have indicated that circRNAs participate in the initiation and development of various cancers such as breast cancer. Breast cancer is a heterogeneous disease, which is the most common malignancy in women. The incidence and mortality rates of breast cancer indicate that it is the leading cause of cancer-related deaths. The goal of the present review is to introduce biogenesis, function characteristics and types of circRNAs, and also their biological functions on breast cancer, especially as miRNA sponges. Additionally, we discuss their use as a new therapeutic target for the treatment of breast cancer.





Similar content being viewed by others
Abbreviations
- BCa:
-
Breast cancer
- ncRNA:
-
Non-coding RNA
- miRNA:
-
microRNA
- circRNA:
-
Circular RNA,
- RBPs:
-
RNA-binding proteins
- MER:
-
miRNA response element
- GFRA1:
-
GDNF family receptor alpha 1 protein
- ARHGEF28:
-
Rho guanine nucleotide exchange factor 28
- ZEB:
-
Zinc finger E-box-binding homeobox 1
- ETS1:
-
ETS proto-oncogene 1
- TFF1:
-
Trefoil factor 1
- ZNF83:
-
Zinc finger protein 83
- USF1:
-
Upstream transcription factor 1
- BCL11A:
-
B-cell lymphoma/leukemia 11A
- SOCS3:
-
Suppressor of cytokine signaling 3
- RAF1:
-
RAF 1 proto-oncogene serine/threonine-protein kinase
- NTRK3:
-
Neurotrophic receptor tyrosine kinase 3
- FOXC1:
-
Forkhead box C1
- ICTH:
-
Itchy E3 ubiquitin protein ligase
- MTA1:
-
Metastasis-associated protein 1
- E2F3:
-
E2F transcription factor 3
- MUC19:
-
Mucin 19, oligomeric
- CBX8:
-
Chromobox 8
- Notch1:
-
Notch receptor 1
- CCNE1:
-
Cyclin E1
- RFC3:
-
Replication factor C subunit 3
- ASS1:
-
Argininosuccinate synthase 1
- HIPK3:
-
Homeodomain-interacting protein kinase 3
- KIF4A:
-
Kinesin family member 4A
- BARD1:
-
BRCA1-associated RING domain 1
References
Wang X, Fang L. Advances in circular RNAs and their roles in breast cancer. J Exp Clin Cancer Res. 2018;37(1):206.
WHO: Geneva, Switzerland. Breast cancer. http://www.who.int/cancer/prevention/diagnosis-screening/breast-cancer/en/.
Power EJ, Chin ML, Haq MM. Breast cancer incidence and risk reduction in the hispanic population. Cureus. 2018;10:e2235.
Misir S, Aliyazicioglu Y, Demir S, Turan I, Hepokur C. Effect of Turkish propolis on miRNA expression, cell cycle, and apoptosis in human breast cancer (MCF-7) Cells. Nutr Cancer. 2020;72(1):133–45.
Zhou SY, Chen W, Yang SJ, Xu ZH, Hu JH, Zhang HD, et al. The emerging role of circular RNAs in breast cancer. Biosci Rep. 2019;39(6):BSR20190621.
Wijnhoven BPL, Michael MZ, Watson DI. MicroRNAs and cancer. Br J Surg. 2007;94(1):23–30.
Zhang P, Wu W, Chen Q, Xu ZH, Hu JH, Zhang HD, et al. Non-coding RNAs and their integrated networks. Integr Bioinform. 2019;16(3):20190027.
Guil S, Esteller M. RNA-RNA interactions in gene regulation: the coding and noncoding players. Trends Biochem Sci. 2015;40(5):248–56.
Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer. 2019;18(1):6.
Klinge CM. Non-coding RNAs in breast cancer: intracellular and intercellular communication. Noncoding RNA. 2018;4(4):40.
Kazimierczyk M, Kasprowicz MK, Kasprzyk ME, Wrzesinski J. Human long noncoding RNA interactome: detection, characterization and function. Int J Mol Sci. 2020;21(3):1027.
Zhao W, Dong M, Pan J, Wang Y, Zhou J, Ma J, et al. Circular RNAs: a novel target among non–coding RNAs with potential roles in malignant tumors (Review). Mol Med Rep. 2019;20(4):3463–74.
Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94.
Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18:116.
Li M, Ding W, Sun T, Tariq MA, Xu T, Li P, et al. Biogenesis of circular RNAs and their roles in cardiovascular development and pathology. FEBS J. 2018;285(2):220–32.
Floris G, Zhang L, Follesa P, Sun T. Regulatory role of circular RNAs and neurological disorders. Mol Neurobiol. 2017;54(7):5156–65.
Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215.
Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64.
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N6-methyladenosine. Cell Res. 2017;27(5):626–41.
Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE. 2012;7:e30733.
Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–8.
Celotto AM, Graveley BR. Alternative splicing of the Drosophila Dscam pre-mRNA is both temporally and spatially regulated. Genetics. 2001;159:599–608.
Shabaninejad Z, Vafadar A, Movahedpour A, Ghasemi Y, Namdar A, Fathizadeh H, et al. CircularRNAs in cancer: new insights into functions and implications in ovarian cancer. J Ovarian Res. 2019;12:84.
Kim SJ, Oh JS, Shin JY, Lee KD, Sung KW, et al. Development of microRNA-145 for therapeutic application in breast cancer. J Control Release. 2011;155:427–34.
Hogg DR, Harries LW. Human genetic variation and its effect on miRNA biogenesis, activity and function. Biochem Soc Trans. 2014;42:1184–9.
Mostafapour KH, Salimi M, Khori V, Rastkari N, Amanzadeh A, Salimi M. Mitochondrial apoptosis ınduced by chamaemelum nobile extract in breast cancer cells. Iran J Pharm Res. 2016;15:197–204.
Verduci L, Strano S, Yarden Y, Blandino G. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–80.
Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–86.
Chen Y, Varani G. Engineering RNA-binding proteins for biology. FEBS J. 2013;280(16):3734–54.
Greene J, Baird AM, Brady L, Lim M, Gray SG, McDermott R, et al. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38.
Du WW, Yang WN, Chen Y, Wu ZK, Foster FS, Yang ZG, et al. Foxo3 circular RNA promotes cardiac senescence by modulating multiple factors associated with stress and senescence responses. Eur Heart J. 2017;38:1402–12.
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58.
Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66.
Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017;14(3):361–9.
Turner M, Galloway A, Vigorito E. Noncoding RNA and its associated proteins as regulatory elements of the immune system. Nat Immunol. 2014;15:484–4491.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, et al. Circular intronic long noncoding RNAs. Mol Cell. 2013;51(6):792–806.
He RF, Liu P, Xie XM, Zhou YJ, Liao QJ, Xiong W, et al. circGFRA1 and GFRA1 act as ceRNAs in triple negative breast cancer by regulating miR-34a. J Exp Clin Cancer Res. 2017;36:145.
Burd CE, Jeck WR, Liu Y, Sanoff HK, Wang Z, Sharpless NE. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233.
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–91.
Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of circRNAs. Mol Cell. 2017;66(9–21):e27.
Yang Y, Gao X, Zhang M, Yan S, Sun C, Xiao F, et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J Natl Cancer Inst. 2018;110:304–15.
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–14.
Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat Commun. 2018;9:4475.
Li L, Yuan L, Luo J, Gao J, Guo J, Xie X. MiR-34a inhibits proliferation and migration of breast cancer through down-regulation of Bcl-2 and SIRT1. Clin Exp Med. 2013;13(2):109–17.
Liang HF, Zhang XZ, Liu B, Jia GT, Li WL. Circular RNA circ-ABCB10 promotes breast cancer proliferation and progression through sponging miR-1271. Am J Cancer Res. 2017;7(7):1566–76.
Tang YY, Zhao P, Zou TN, Duan JJ, Zhi R, Yang SY, et al. Circular RNA hsa_circ_0001982 promotes breast cancer cell carcinogenesis through decreasing miR-143. DNA Cell Biol. 2017;36:11.
Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10(9):1229–42.
Shukla K, Sharma AK, Ward A, Will R, Hielscher T, Balwierz A, et al. MicroRNA-30c-2-3p negatively regulates NF-kappaB signaling and cell cycle progression through downregulation of TRADD and CCNE1 in breast cancer. Mol. Oncol. 2015;9(6):1106–19.
Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10(2):55.
Korpal M, Kang Y. The emerging role of miR-200 family of microRNAs in epithelial-mesenchymal transition and cancer metastasis. RNA Biol. 2008;5:115–9.
Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603.
Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720.
Zhang HD, Jiang LH, Hou JC, Zhong SL, Zhou SY, Zhu LP, et al. Circular RNA hsa_circ_0052112 promotes cell migration and invasion by acting as sponge for miR-125a-5p in breast cancer. Biomed Pharmacother. 2018;107:1342–53.
Jiang L, Huang Q, Zhang S, Zhang Q, Chang J, Qiu X, et al. Hsa-miR-125a- 3p and hsa-miR-125a-5p are downregulated in non-small cell lung cancer and have inverse effects on invasion and migration of lung cancer cells. BMC Cancer. 2010;10:318.
Zeng K, He B, Yang BB, Xu T, Chen X, Xu M, et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17(1):160.
Zhao J, Zou H, Han C, Ma J, Zhao J, Tang J. Circlular RNA BARD1 (hsa_circ_0001098) overexpression in breast cancer cells with TCDD treatment could promote cell apoptosis via miR-3942/BARD1 axis. Cell Cycle. 2018;17:2731–44.
Yuan C, Zhou L, Zhang L, Yin K, Peng J, Sha R, et al. Identification and integrated analysis of key differentially expressed circular RNAs in ER-positive subtype breast cancer. Epigenomics. 2019;11:297–321.
Tang H, Huang X, Wang J, Yang L, Kong Y, Gao G, et al. circKIF4A acts as a prognostic factor and mediator to regulate the progression of triple-negative breast cancer. Mol Cancer. 2019;18(1):23.
Kong Y, Yang L, Wei W, Lyu N, Zou Y, Gao G, et al. CircPLK1 sponges miR-296-5p to facilitate triple-negative breast cancer progression. Epigenomics. 2019;11(10):1163–76.
Hou JC, Xu Z, Zhong SL, Zhang H, Jiang LH, Chen X, et al. Circular RNA circASS1 Is downregulated in breast cancer cells MDA-MB-231 and suppressed invasion and migration. Epigenomics. 2019;11(2):199–213.
Hu Y, Guo F, Zhu H, Tan X, Zhu X, Liu X, et al. Circular RNA-0001283 suppresses breast cancer proliferation and invasion via MiR-187/HIPK3 axis. Med Sci Monit. 2020;26:e921502-1–9.
Chen B, Wei W, Huang X, Xie X, Kong Y, Dai D, et al. circEPSTI1 as a prognostic marker and mediator of triple-negative breast cancer progression. Theranostics. 2018;8(14):4003–15.
Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175.
Gao D, Qi X, Zhang X, Fang K, Guo Z, Li L. hsa_circRNA_0006528 as a competing endogenous RNA promotes human breast cancer progression by sponging miR-7-5p and activating the MAPK/ERK signaling pathway. Mol Carcinog. 2019;58(4):554–64.
Shi Y, Luo X, Li P, Tan J, Wang X, Xiang T, et al. miR-7-5p suppresses cell proliferation and induces apoptosis of breast cancer cells mainly by targeting REGγ. Cancer Lett. 2015;358:27–36.
Yao Y, Zhao Z, Han X, Lin S, Nie C, Fang H. Circular RNA-100219 promotes breast cancer progression by binding to microRNA-485-3p. J BUON. 2019;24(2):501–8.
Wu J, Jiang Z, Chen C, Hu Q, Fu Z, Chen J, et al. CircIRAK3 sponges miR-3607 to facilitate breast cancer metastasis. Cancer Lett. 2018;430:179–92.
Wang ST, Liu LB, Li XM, Wang YF, Xie PJ, Li Q, et al. Circ-ITCH regulates triple-negative breast cancer progression through the Wnt/β-catenin pathway. Neoplasma. 2019;66(2):232–9.
Wang S, Li Q, Wang Y, Li X, Wang R, Kang Y, Xue X, Meng R, Wei Q, Feng X. Upregulation of circ-UBAP2 predicts poor prognosis and promotes triple-negative breast cancer progression through the miR-661/MTA1 pathway. Biochem Biophys Res Commun. 2018;505(4):996–1002.
Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–63.
Song L, Xiao Y. Downregulation of hsa_circ_0007534 suppresses breast cancer cell proliferation and invasion by targeting miR-593/MUC19 signal pathway. Biochem Biophys Res Commun. 2018;503(4):2603–10.
Xu Y, Yao Y, Leng K, Ji D, Qu L, Liu Y, et al. Increased expression of circular RNA circ_0005230 indicates dismal prognosis in breast cancer and regulates cell proliferation and invasion via miR-618/CBX8 signal pathway. Cell Physiol Biochem. 2018;51(4):1710–22.
Yan L, Zheng M, Wang H. Circular RNA hsa_circ_0072309 inhibits proliferation and invasion of breast cancer cells via targeting miR-492. Cancer Manag Res. 2019;11:1033–41.
Wang H, Xiao Y, Wu L, Ma D. Comprehensive circular RNA profiling reveals the regulatory role of the circRNA-000911/miR-449a pathway in breast carcinogenesis. Int J Oncol. 2018;52(3):743–54.
Yang R, Xing L, Zheng X, Sun Y, Wang X, Chen J. The circRNA circAGFG1 acts as a sponge of miR-195-5p to promote triple-negative breast cancer progression through regulating CCNE1 expression. Mol Cancer. 2019;18(1):4.
Zhou J, Zhang WW, Peng F, Sun JY, He ZY, Wu SG. Downregulation of hsa_circ_0011946 suppresses the migration and invasion of the breast cancer cell line MCF-7 by targeting RFC3. Cancer Manag Res. 2018;10:535–44.
Geng Y, Jiang J, Wu C. Function and clinical significance of circRNAs in solid tumors. J Hematol Oncol. 2018;11:98.
Zhang M, Xin Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J Hematol Oncol. 2018;11:21.
Puttaraju M, Been MD. Group I permuted intron-exon (PIE) sequences selfsplice to produce circular exons. Nucleic Acids Res. 1992;20(20):5357–64.
Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang JL, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15(3):611–24.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
About this article
Cite this article
Misir, S., Hepokur, C., Aliyazicioglu, Y. et al. Circular RNAs serve as miRNA sponges in breast cancer. Breast Cancer 27, 1048–1057 (2020). https://doi.org/10.1007/s12282-020-01140-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-020-01140-w