Abstract
Background
The genotype of Fanconi Anemia complementation group M (FANCM) was previously found to be associated with breast cancer risk in several populations. Here, we studied the expression of FANCM and its correlation with clinical characteristics in Chinese patients with breast cancer.
Methods
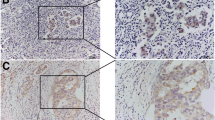
We performed an immunohistochemical study of FANCM protein in clinical breast cancer tissues from 310 patients along with 44 adjacent tissues.
Results
FANCM protein level is lower in triple-negative breast cancer tissues than in other subtypes (P = 0.008). In addition, high FANCM expression correlated with pathology type IDC (P = 0.040), estrogen receptor positive (P < 0.001), progesterone receptor positive (P = 0.001), and low Ki-67 status (P = 0.003). Multivariate analysis revealed that FANCM status was an independent prognostic factor for overall survival (P = 0.017) in luminal B breast cancer.
Conclusions
FANCM levels are significantly associated with different subtypes of human breast cancer. Specifically, FANCM could play a role in the progression of luminal B breast cancer.



Similar content being viewed by others
References
Siegel R, Miller K, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
Chen W, Zheng R, Baade P, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China. CA Cancer J Clin. 2015;66(2):115–32.
Heilmann K. Epigenetic characterization of the C3(1) SV40T mouse model of human breast cancer. 2017.
Holm J, Eriksson L, Ploner A, Eriksson M, Rantalainen M, Li J, et al. Assessment of breast cancer risk factors reveals subtype heterogeneity. Cancer Res. 2017;77(13):3708–17.
Martín-Sánchez E, Mendaza S, Ulazia-Garmendia A, Monreal-Santesteban I, Córdoba A, Vicente-García F, et al. CDH22hypermethylation is an independent prognostic biomarker in breast cancer. Clin Epigenetics. 2017;9(1):7.
Ling C, Huang J, Yan Z, Li Y, Ohzeki M, Ishiai M, et al. Bloom syndrome complex promotes FANCM recruitment to stalled replication forks and facilitates both repair and traverse of DNA interstrand crosslinks. Cell Discov. 2016;2:16047.
Ali AM, Singh TR, Meetei AR. FANCM-FAAP24 and FANCJ: FA proteins that metabolize DNA. Mutat Res. 2009;668(1–2):20–6.
Xue X, Sung P, Zhao X. Functions and regulation of the multitasking FANCM family of DNA motor proteins. Genes Dev. 2015;29(17):1777–88.
Rohleder F, Huang J, Xue Y, Kuper J, Round A, Seidman M, et al. FANCM interacts with PCNA to promote replication traverse of DNA interstrand crosslinks. Nucl Acids Res. 2016;44(7):3219–32.
Dicks E, Song H, Ramus SJ, Oudenhove EV, Tyrer JP, Intermaggio MP, et al. Germline whole exome sequencing and large-scale replication identifies FANCM as a likely high grade serous ovarian cancer susceptibility gene. Oncotarget. 2017;8(31):50930–40.
Neidhardt G, Hauke J, Ramser J, Gross E, Gehrig A, Muller CR, et al. Association between loss-of-function mutations within the FANCM gene and early-onset familial breast cancer. JAMA Oncol. 2017;3(9):1245–8.
Catucci I, Osorio A, Arver B, Neidhardt G, Bogliolo M, Zanardi F, et al. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet Med. 2017. https://doi.org/10.1038/gim.2017.123.
Kiiski JI, Fagerholm R, Tervasmaki A, Pelttari LM, Khan S, Jamshidi M, et al. FANCM c.5101C > T mutation associates with breast cancer survival and treatment outcome. Int J Cancer. 2016;139(12):2760–70.
Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013;24(9):2206–23.
Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013;31(31):3997–4013.
Peterlongo P, Catucci I, Colombo M, Caleca L, Mucaki E, Bogliolo M, et al. FANCM c.5791C > T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity and is a familial breast cancer risk factor. Hum Mol Genet. 2015;24(18):5345–55.
Rashid MU, Muhammad N, Khan FA, Hamann U. Absence of the FANCM c.5101C > T mutation in BRCA1/2-negative triple-negative breast cancer patients from Pakistan. Breast Cancer Res Treat. 2015;152(1):229–30.
Kiiski JI, Pelttari LM, Khan S, Freysteinsdottir ES, Reynisdottir I, Hart SN, et al. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc Natl Acad Sci USA. 2014;111(42):15172–7.
Kiiski JI, Tervasmaki A, Pelttari LM, Khan S, Mantere T, Pylkas K, et al. FANCM mutation c.5791C > T is a risk factor for triple-negative breast cancer in the Finnish population. Breast Cancer Res Treat. 2017;66(1):217–26.
Pan X, Drosopoulos WC, Sethi L, Madireddy A, Schildkraut CL, Zhang D. FANCM, BRCA1, and BLM cooperatively resolve the replication stress at the ALT telomeres. Proc Natl Acad Sci USA. 2017;114(29):E5940–9.
Krammer J, Pinker-Domenig K, Robson ME, Gonen M, Bernard-Davila B, Morris EA, et al. Breast cancer detection and tumor characteristics in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res Treat. 2017;163(3):565–71.
Wong-Brown MW, Meldrum CJ, Carpenter JE, Clarke CL, Narod SA, Jakubowska A, et al. Prevalence of BRCA1 and BRCA2 germline mutations in patients with triple-negative breast cancer. Breast Cancer Res Treat. 2015;150(1):71–80.
Sinn HP, Schneeweiss A, Keller M, Schlombs K, Laible M, Seitz J, et al. Comparison of immunohistochemistry with PCR for assessment of ER, PR, and Ki-67 and prediction of pathological complete response in breast cancer. BMC Cancer. 2017;17(1):124.
Braunstein LZ, Taghian AG, Niemierko A, Salama L, Capuco A, Bellon JR, et al. Breast-cancer subtype, age, and lymph node status as predictors of local recurrence following breast-conserving therapy. Breast Cancer Res Treat. 2017;161(1):173–9.
Cheang MCU, Chia SK, Voduc D, Gao DX, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101(10):736–50.
Bustreo S, Osella-Abate S, Cassoni P, Donadio M, Airoldi M, Pedani F, et al. Optimal Ki67 cut-off for luminal breast cancer prognostic evaluation: a large case series study with a long-term follow-up. Breast Cancer Res Treat. 2016;157(2):363–71.
Berton S, Cusan M, Segatto I, Citron F, D’Andrea S, Benevol S, et al. Loss of p27 kip1 increases genomic instability and induces radio-resistance in luminal breast cancer cells. Sci Rep. 2017;7(1):595.
Schwab RA, Blackford AN, Niedzwiedz W. ATR activation and replication fork restart are defective in FANCM-deficient cells. EMBO J. 2010;29(4):806–18.
Funding
This work was supported by the National Natural Science Foundation of China (81572591).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared no conflicts of interest.
Ethical approval
Ethical approval is in accordance with the Declaration of Helsinki.
About this article
Cite this article
Wang, Y., Wang, J., Long, F. et al. Correlation of FANCM expression with clinical factors in luminal B breast cancer. Breast Cancer 25, 431–437 (2018). https://doi.org/10.1007/s12282-018-0841-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-018-0841-0