Abstract
Background
Breast cancer (BC) is the most prevalent cancer and the main cause of cancer deaths among females around the world. For early diagnosis of BC, there would be an immediate and essential requirement to search for sensitive biomarkers.
Methods
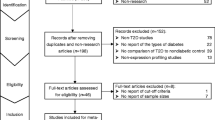
To identify candidate miRNA biomarkers for BC, we performed a general systematic review regarding the published miRNA profiling researches comparing miRNA expression level between BC and normal tissues. A miRNA ranking system was selected, which considered frequency of comparisons in direction and agreement of differential expression.
Results
We determined that two miRNAs (mir-21 and miR-210) were upregulated consistently and six miRNAs (miR-145, miR-139-5p, miR-195, miR-99a, miR-497 and miR-205) were downregulated consistently in at least three studies. MiR-21 as the most consistently reported miRNA was upregulated in six profiling studies.
Conclusions
Although these miRNAs require being validated and further investigated, they could be potential candidates for BC miRNA biomarkers and used for early prognosis or diagnosis.

Similar content being viewed by others
References
Torre Lindsey A, Siegel Rebecca L, Ferlay Jacques, Lortet-Tieulent Joannie, Jemal Ahmedin. Global Cancer Statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Smigal CJA, Ward E, Cokkinides V, Smith R, Howe HL, et al. Trends in breast cancer by race and ethnicity: update 2006. CA Cancer J Clin. 2006;56:168–83.
Tryggvadottir LGM, Bray F, Klint A, Hakulinen T, Storm HH, et al. Trends in the survival of patients diagnosed with breast cancer in the Nordic countries 1964–2003 followed up to the end of 2006. Acta Oncol. 2010;49:624–31.
Rosso SGA, Zanetti R, Bray F, Zakelj M, Zagar T, et al. Up-to-date estimates of breast cancer survival for the years 2000–2004 in 11 European countries: the role of screening and a comparison with data from the United States. Eur J Cancer. 2010;46:3351–7.
Holleczek BAV, Stegmaier C, Brenner H. Trends in breast cancer survival in Germany from 1976 to 2008—a period analysis by age and stage. Cancer Epidemiol. 2011;35:399–406.
Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER cancer statistics review, 1975-2008. National Cancer Institute, Bethesda, MD. Based on November 2010 SEER data submission, posted to the SEER web site, 2011. http://seer.cancer.gov/csr/1975_2008/. Accessed 1 Aug 2017
Sankaranarayanan RSR, Brenner H, Chen K, Chia KS, Chen JG, et al. Cancer survival in Africa, Asia, and Central America: a population-based study. Lancet Oncol. 2010;11:165–73.
Wiemer EA. The role of microRNAs in cancer: no small matter. Eur J Cancer Epidemiol. 2007;43:1529–44.
He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31.
Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5.
Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–97.
Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–7.
Kent OA, Mendell JT. A small piece in the cancer puzzle: microRNAs as tumor suppressors and oncogenes. Oncogene. 2006;25:6188–96.
Iorio MVCC. MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med. 2012;4:143–59.
Ma YZP, Yang J, Liu Z, Yang Z, et al. Candidate microRNA biomarkers in human colorectal cancer: systematic review profiling studies and experimental validation. Int J Cancer. 2012;130:2077–87.
Chan SKGO, Tai IT, Jones SJ. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol Biomark Prev. 2008;17:543–52.
Griffith OLMA, Jones SJ, Wiseman SM. Meta-analysis and meta-review of thyroid cancer gene expression profiling studies identifies important diagnostic biomarkers. J Clin Oncol. 2006;24:5043–51.
Ji-Lin Wang YH, Xuan Kong, Zhen-Hua Wang, Hao-Yan Chen, Jie Xu, Jing-Yuan Fang. Candidate microRNA biomarkers in human gastric cancer: a systematic review and validation study. PLoS One. 2013;8:1–8.
Sun J, Li M, Li Z, Xue J, Lan X, Zhang C, et al. Identification and profiling of conserved and novel microRNAs from Chinese Qinchuan bovine longissimus thoracis. BMC Genom. 2013;14:42.
Hu J, Xu J, Wu Y, Chen Q, Zheng W, Lu X, et al. Identification of microRNA-93 as a functional dysregulated miRNA in triple-negative breast cancer. Tumour Biol. 2015;36:251–8.
Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–70.
Matamala N, Vargas MT, Gonzalez-Campora R, Minambres R, Arias JI, Menendez P, et al. Tumor microRNA expression profiling identifies circulating microRNAs for early breast cancer detection. Clin Chem. 2015;61:1098–106.
Navon R, Wang H, Steinfeld I, Tsalenko A, Ben-Dor A, Yakhini Z. Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One. 2009;4:e8003.
Pena-Chilet M, Martinez MT, Perez-Fidalgo JA, Peiro-Chova L, Oltra SS, Tormo E, et al. MicroRNA profile in very young women with breast cancer. BMC Cancer. 2014;14:529.
Sempere LF, Christensen M, Silahtaroglu A, Bak M, Heath CV, Schwartz G, et al. Altered MicroRNA expression confined to specific epithelial cell subpopulations in breast cancer. Cancer Res. 2007;67:11612–20.
Sun EH, Zhou Q, Liu KS, Wei W, Wang CM, Liu XF, et al. Screening miRNAs related to different subtypes of breast cancer with miRNAs microarray. Eur Rev Med Pharmacol Sci. 2014;18:2783–8.
Yan LX, Huang XF, Shao Q, Huang MY, Deng L, Wu QL, et al. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–60.
Zhang K, Zhao S, Wang Q, Yang HS, Zhu J, Ma R. Identification of microRNAs in nipple discharge as potential diagnostic biomarkers for breast cancer. Ann Surg Oncol. 2015;22(Suppl 3):S536–44.
Zhang M, Liu D, Li W, Wu X, Gao C, Li X. Identification of featured biomarkers in breast cancer with microRNA microarray. Arch Gynecol Obstet. 2016;294:1047–53.
Bhattacharya A, Cui Y. SomamiR 2.0: a database of cancer somatic mutations altering microRNA–ceRNA interactions. Nucleic Acids Res. 2016;44(D1):D1005–10.
Meng X, Wang J, Yuan C, Li X, Zhou Y, Hofestadt R, et al. CancerNet: a database for decoding multilevel molecular interactions across diverse cancer types. Oncogenesis. 2015;4:e177.
Thakur S, Grover RK, Gupta S, Yadav AK, Das BC. Identification of specific miRNA signature in paired sera and tissue samples of indian women with triple negative breast cancer. PLoS One. 2016;11:e0158946.
Kodahl AR, Lyng MB, Binder H, Cold S, Gravgaard K, Knoop AS, et al. Novel circulating microRNA signature as a potential non-invasive multi-marker test in ER-positive early-stage breast cancer: a case control study. Mol Oncol. 2014;8:874–83.
Shimomura A, Shiino S, Kawauchi J, Takizawa S, Sakamoto H, Matsuzaki J, et al. Novel combination of serum microRNA for detecting breast cancer in the early stage. Cancer Sci. 2016;107:326–34.
Alder HTC, Chen H, Jiang Y, Smalley KJ, et al. Dysregulation of miR-31 and miR-21 induced by zinc deficiency promotes esophageal cancer. Carcinogenesis. 2012;33:1736–44.
Iliopoulos DJS, Hirsch HA, Bulyk ML, Struhl K. STAT3 activation of miR-21 and miR-181b-1 via PTEN and CYLD are part of the epigenetic switch linking inflammation to cancer. Mol Cell. 2010;39:493–506.
Han MWY, Liu M, Bi X, Bao J, et al. MiR-21 regulates epithelial-mesenchymal transition phenotype and hypoxia-inducible factor-1 a expression in third-sphere forming breast cancer stem cell-like cells. Cancer Sci. 2012;103:1058–64.
Fang HXJ, Zhang M, Zhao Z, Wan Y, Yao Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9:953–61.
Tang YZX, Ji J, Chen L, Cao J, Luo J, Zhang S. High expression levels of miR-21 and miR-210 predict unfavorable survival in breast cancer: a systemic review and meta-analysis. Int J Biol Markers. 2015;30:e347–58.
Liu TY, HZ, Du SM, Li J, Wen XH. Expression of microRNA-210 in tissue and serum of renal carcinoma patients and its effect on renal carcinoma cell proliferation, apoptosis, and invasion. Genet Mol Res. 2016;15:15017746.
Rothé FIM, Chaboteaux C, Haibe-Kains B, Kheddoumi N, Majjaj S, Badran B, Fayyad-Kazan H, Desmedt C, Harris AL, Piccart M, Sotiriou C. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS One. 2011;6:e20980.
Liu DXH, Wang F, Chen C, Long J. MicroRNA-210 interacts with FBXO31 to regulate cancer proliferation cell cycle and migration in human breast cancer. Onco Targets Ther. 2016;23:5245–55.
Bandrés ECE, Agirre X, Malumbres R, Zárate R, Ramirez N, Abajo A, Navarro A, Moreno I, Monzó M, García-Foncillas J. Identification by Real-time PCR of 13 mature microRNAs differentially expressed in colorectal cancer and non-tumoral tissues. Mol Cancer. 2006;19(5):29–39.
Yanaihara NCN, Bowman E, Seike M, Kumamoto K, Yi M, Stephens RM, Okamoto A, Yokota J, Tanaka T, Calin GA, Liu CG, Croce CM, Harris CC. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–98.
LE Szczyrba J, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stöhr R, Hartmann A, Wullich B, Grässer F. The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res. 2010;8:529–38.
Kano MSN, Kikkawa N, Fujimura L, Hoshino I, Akutsu Y, Chiyomaru T, Enokida H, Nakagawa M, Matsubara H. miR-145, miR-133a and miR-133b: tumor suppressive miRNAs target FSCN1 in esophageal squamous cell carcinoma. Int J Cancer Cell. 2010;127:2804–14.
Zhao HKX, Xia X, Wo L, Gu X, Hu Y, Xie X, Chang H, Lou L, Shen X. miR-145 suppresses breast cancer cell migration by targeting FSCN-1 and inhibiting epithelial-mesenchymal transition. Am J Transl Res. 2016;8:3106–14.
Ding YZC, Zhang J, Zhang N, Li T, Fang J, Zhang Y, Zuo F, Tao Z, Tang S, Zhu W, Chen H, Sun X. miR-145 inhibits proliferation and migration of breast cancer cells by directly or indirectly regulating TGF-β1 expression. Int J Oncol. 2017;50:1701–10.
Enders KO, Ng RL, Vivian Shin Y, Hong Chuan Jin, Candy Leung PH, Edmond Ma SK, Roberta Pang, Daniel Chua, Kent-Man Chu, Law WL, Simon Law YK, Ronnie Poon TP, Ava Kwong. Circulating microRNAs as specific biomarkers for breast cancer detection. PLoS One. 2013;8:e53141.
Chengcao Sun MS, Shujun Li, Xiaodong Sun, Cuili Yang, Yongyong Xi, Liang Wang, Feng Zhang, Yongyi Bi, Yunfeng Fu, Dejia Li. Hsa-miR-139-5p inhibits proliferation and causes apoptosis associated with down-regulation of c-Met. Oncotarget. 2015;6:39756–92.
Yonemori MSN, Yoshino H, Matsushita R, Miyamoto K, Nakagawa M, Enokida H. Dual tumor-suppressors miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in bladder cancer. Cancer Sci. 2016;107:1233–42.
Zhou QHL, Zhou YX, Li Y. MiR-195 suppresses cervical cancer migration and invasion through targeting Smad3. Int J Gynecol Cancer. 2016;26:817–24.
Zhang XXJ, Jiang T, Liu G, Wang D, Lu Y. MicroRNA-195 suppresses colorectal cancer cells proliferation via targeting FGF2 and regulating Wnt/β-catenin pathway. Am J Cancer Res. 2016;6:2631–40.
Srivastava AGH, Dimtchev A, Ramalinga M, Chijioke J, Marian C, Oermann EK, Uhm S, Kim JS, Chen LN, Li X, Berry DL, Kallakury BV, Chauhan SC, Collins SP, Suy S, Kumar D. MicroRNA profiling in prostate cancer–the diagnostic potential of urinary miR-205 and miR-214. PLoS One. 2013;8:e76994.
Salajegheh AVH, Rahman Md A, Amin M, Smith RA, Lam AK. Modulatory role of miR-205 in angiogenesis and progression of thyroid cancer. J Mol Endocrinol. 2015;55:183–96.
Wang PMX, Huang Y, Lv Z, Liu J, Wang G, Meng W, Xue S, Zhang Q, Zhang P, Chen G. MicroRNA-497 inhibits thyroid cancer tumor growth and invasion by suppressing BDNF. Oncotarget. 2017;8:2825–34.
Zhang YZZ, Li Z, Gong D, Zhan B, Man X, Kong C. MicroRNA-497 inhibits the proliferation, migration and invasion of human bladder transitional cell carcinoma cells by targeting E2F3. Oncol Rep. 2016;36:1293–300.
Feng Y KY, He Y, Liu J, Liang B, Yang P, Yu Z. MicroRNA-99a acts as a tumor suppressor and is down-regulated in bladder cancer. BMC Urol. 14 2014
Yu SHZC, Dong FS, Zhang YM. miR-99a suppresses the metastasis of human non-small cell lung cancer cells by targeting AKT1 signaling pathway. J Cell Biochem. 2015;116:268–76.
Huang HGLX, Wu S, Jian B. MiR-99a inhibits cell proliferation and tumorigenesis through targeting mTOR in human anaplastic thyroid cancer. Asian Pac J Cancer Prev. 2015;16:4937–44.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
About this article
Cite this article
Adhami, M., Haghdoost, A.A., Sadeghi, B. et al. Candidate miRNAs in human breast cancer biomarkers: a systematic review. Breast Cancer 25, 198–205 (2018). https://doi.org/10.1007/s12282-017-0814-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12282-017-0814-8