Abstract
Purpose of Review
The purpose of this study is to analyze the current knowledge about matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS) identification of clinical yeasts comparing with the ribosomal DNA sequencing identification. We also review the lasts taxonomical and nomenclature changes of clinical yeasts.
Recent Findings
In most studies, the two main commercially available, MALDI Biotyper and VITEK MS, correctly identified more than 90% of yeasts isolates. Most drawbacks are due to low discrimination between closely related species or due to a lack of the reference main spectra (MSP) in the database. The addition of MSPs in the databases improves the performance.
Summary
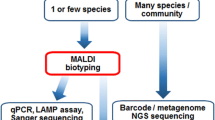
MALDI-TOF MS has proven to be good for the identification of clinical yeasts and to differentiate some closely related species or species undifferentiated by conventional techniques. However, some species remain difficult to differentiate by this technique and should be identified by ribosomal DNA sequencing. Accurate and rapid yeasts identification could improve the management of patients with yeasts infections.
Similar content being viewed by others
References
Kurtzman CP, Fell JW, Boekhout T. The yeasts, a taxonomic study. London: Elsevier; 2011.
Alcoba-Flórez J, Méndez-Alvarez S, Cano J, Guarro J, Pérez-Roth E, del Pilar Arévalo M. Phenotypic and molecular characterization of Candida nivariensis sp. nov., a possible new opportunistic fungus. J Clin Microbiol. 2005;43(8):4107–11.
Correia A, Sampaio P, James S, Pais C. Candida bracarensis sp. nov., a novel anamorphic yeast species phenotypically similar to Candida glabrata. Int J Syst Evol Microbiol. 2006;56(Pt 1):313–7.
Sullivan DJ, Westerneng TJ, Haynes KA, Bennett DE, Coleman DC. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiol Read Engl. 1995;141(Pt 7):1507–21.
Tavanti A, Davidson AD, Gow NAR, Maiden MCJ, Odds FC. Candida orthopsilosis and Candida metapsilosis spp. nov. to replace Candida parapsilosis groups II and III. J Clin Microbiol. 2005;43(1):284–92.
Tietz HJ, Hopp M, Schmalreck A, Sterry W, Czaika V. Candida africana sp. nov., a new human pathogen or a variant of Candida albicans? Mycoses. 2001;44(11–12):437–45.
Vaughan-Martini A, Kurtzman CP, Meyer SA, O’Neill EB. Two new species in the Pichia guilliermondii clade: Pichia caribbica sp. nov., the ascosporic state of Candida fermentati, and Candida carpophila comb. nov. FEMS Yeast Res. 2005;5(4–5):463–9.
Cendejas-Bueno E, Kolecka A, Alastruey-Izquierdo A, Theelen B, Groenewald M, Kostrzewa M, et al. Reclassification of the Candida haemulonii complex as Candida haemulonii (C. haemulonii group I), C. duobushaemulonii sp. nov. (C. haemulonii group II), and C. haemulonii var. vulnera var. nov.: three multiresistant human pathogenic yeasts. J Clin Microbiol. 2012;50(11):3641–51.
Chaves GM, Terçarioli GR, Padovan ACB, Rosas RC, Ferreira RC, Melo ASA, et al. Candida mesorugosa sp. nov., a novel yeast species similar to Candida rugosa, isolated from a tertiary hospital in Brazil. Med Mycol. 2013;51(3):231–42.
Paredes K, Sutton DA, Cano J, Fothergill AW, Lawhon SD, Zhang S, et al. Molecular identification and antifungal susceptibility testing of clinical isolates of the Candida rugosa species complex and proposal of the new species Candida neorugosa. J Clin Microbiol. 2012;50(7):2397–403.
Groenewald M, Daniel H-M, Robert V, Poot GA, Smith MT. Polyphasic re-examination of Debaryomyces hansenii strains and reinstatement of D. hansenii, D. fabryi and D. subglobosus. Persoonia. 2008;21:17–27.
Castanheira M, Woosley LN, Diekema DJ, Jones RN, Pfaller MA. Candida guilliermondii and other species of candida misidentified as Candida famata: assessment by vitek 2, DNA sequencing analysis, and matrix-assisted laser desorption ionization-time of flight mass spectrometry in two global antifungal surveillance programs. J Clin Microbiol. 2013;51(1):117–24.
Desnos-Ollivier M, Ragon M, Robert V, Raoux D, Gantier J-C, Dromer F. Debaryomyces hansenii (Candida famata), a rare human fungal pathogen often misidentified as Pichia guilliermondii (Candida guilliermondii). J Clin Microbiol. 2008;46(10):3237–42.
Kathuria S, Singh PK, Sharma C, Prakash A, Masih A, Kumar A, et al. Multidrug-resistant Candida auris misidentified as Candida haemulonii: characterization by matrix-assisted laser desorption ionization-time of flight mass spectrometry and DNA sequencing and its antifungal susceptibility profile variability by Vitek 2, CLSI broth microdilution, and Etest method. J Clin Microbiol. 2015;53(6):1823–30.
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, et al. The amsterdam declaration on fungal nomenclature. IMA Fungus. 2011;2(1):105–12.
Taylor JW. One fungus = one name: DNA and fungal nomenclature twenty years after PCR. IMA Fungus. 2011;2(2):113–20.
McNeill J, Barrie F, Buck W, Demoulin V, Greuter W, Hawksworth D, et al. International code of nomenclature for algae, fungi, and plants (Melbourne code). Australia: Koeltz Scientific Books; 2012.
Tsui CKM, Daniel H-M, Robert V, Meyer W. Re-examining the phylogeny of clinically relevant Candida species and allied genera based on multigene analyses. FEMS Yeast Res. 2008;8(4):651–9.
Daniel H-M, Lachance M-A, Kurtzman CP. On the reclassification of species assigned to Candida and other anamorphic ascomycetous yeast genera based on phylogenetic circumscription. Antonie Van Leeuwenhoek. 2014;106(1):67–84.
Lachance MA. In defense of yeast sexual life cycles: the forma asexualis—an informal proposal. Yeast Newletter. 2012;61:24–5.
Hagen F, Khayhan K, Theelen B, Kolecka A, Polacheck I, Sionov E, et al. Recognition of seven species in the Cryptococcus gattii/Cryptococcus neoformans species complex. Fungal Genet Biol FG B. 2015;78:16–48.
Liu X-Z, Wang Q-M, Göker M, Groenewald M, Kachalkin AV, Lumbsch HT, et al. Towards an integrated phylogenetic classification of the Tremellomycetes. Stud Mycol. 2015;81:85–147.
White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols: a guide to methods and applications. New York: Academic Press; 1990. p. 315–22.
Pincus DH, Orenga S, Chatellier S. Yeast identification—past, present, and future methods. Med Mycol. 2007;45(2):97–121.
Iwen PC, Hinrichs SH, Rupp ME. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med Mycol. 2002;40(1):87–109.
Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA, et al. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for fungi. Proc Natl Acad Sci U S A. 2012;109(16):6241–6.
Irinyi L, Serena C, Garcia-Hermoso D, Arabatzis M, Desnos-Ollivier M, Vu D, et al. International Society of Human and Animal Mycology (ISHAM)-ITS reference DNA barcoding database—the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med Mycol. 2015;53(4):313–37.
Fungal XJ. DNA barcoding. Genome. 2016;59(11):913–32.
Taverna CG, Córdoba S, Murisengo OA, Vivot W, Davel G, Bosco-Borgeat ME. Molecular identification, genotyping, and antifungal susceptibility testing of clinically relevant Trichosporon species from Argentina. Med Mycol. 2014;52(4):356–66.
Stielow JB, Lévesque CA, Seifert KA, Meyer W, Iriny L, Smits D, et al. One fungus, which genes? Development and assessment of universal primers for potential secondary fungal DNA barcodes. Persoonia. 2015;35:242–63.
Vu D, Groenewald M, Szöke S, Cardinali G, Eberhardt U, Stielow B, et al. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud Mycol. 2016;85:91–105.
Kurtzman CP, Robnett CJ. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Van Leeuwenhoek. 1998;73(4):331–71.
Fell JW, Boekhout T, Fonseca A, Scorzetti G, Statzell-Tallman A. Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. Int J Syst Evol Microbiol. 2000;50(Pt 3):1351–71.
Taverna CG, Bosco-Borgeat ME, Murisengo OA, Davel G, Boité MC, Cupolillo E, et al. Comparative analyses of classical phenotypic method and ribosomal RNA gene sequencing for identification of medically relevant Candida species. Mem Inst Oswaldo Cruz. 2013;108(2):178–85.
Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J Clin Microbiol. 2002;40(5):1826–30.
Meyer W, Castañeda A, Jackson S, Huynh M, Castañeda E. IberoAmerican Cryptococcal Study Group. Molecular typing of IberoAmerican Cryptococcus neoformans isolates. Emerg Infect Dis. 2003;9(2):189–95.
Croxatto A, Prod’hom G, Greub G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev. 2012;36(2):380–407.
Tan KE, Ellis BC, Lee R, Stamper PD, Zhang SX, Carroll KC. Prospective evaluation of a matrix-assisted laser desorption ionization-time of flight mass spectrometry system in a hospital clinical microbiology laboratory for identification of bacteria and yeasts: a bench-by-bench study for assessing the impact on time to identification and cost-effectiveness. J Clin Microbiol. 2012;50(10):3301–8.
Posteraro B, Vella A, Cogliati M, De Carolis E, Florio AR, Posteraro P, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry-based method for discrimination between molecular types of Cryptococcus neoformans and Cryptococcus gattii. J Clin Microbiol. 2012;50(7):2472–6.
Bader O. Fungal species identification by MALDI-ToF mass spectrometry. Methods Mol Biol Clifton NJ. 2017;1508:323–37.
Santos C, Lima N, Sampaio P, Pais C. Matrix-assisted laser desorption/ionization time-of-flight intact cell mass spectrometry to detect emerging pathogenic Candida species. Diagn Microbiol Infect Dis. 2011;71(3):304–8.
Pfaller MA, Castanheira M. Nosocomial Candidiasis: antifungal stewardship and the importance of rapid diagnosis. Med Mycol. 2016;54(1):1–22.
Santos CR, Francisco E, Mazza M, Padovan ACB, Colombo AL, Lima N. Impact of MALDI-TOF MS in clinical mycology; progress and barriers in diagnostics. In: Shah HR, Gharbia SE, editors. MALDI-TOF and tandem MS for clinical microbiology. Wiley: Chichester; 2017.
Cassagne C, Normand A-C, L’Ollivier C, Ranque S, Piarroux R. Performance of MALDI-TOF MS platforms for fungal identification. Mycoses. 2016;59(11):678–90.
Wang H, Fan Y-Y, Kudinha T, Xu Z-P, Xiao M, Zhang L, et al. A comprehensive evaluation of the Bruker Biotyper MS and Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry systems for identification of yeasts, part of the National China Hospital Invasive Fungal Surveillance Net (CHIF-NET) study, 2012 to 2013. J Clin Microbiol. 2016;54(5):1376–80.
Stefaniuk E, Baraniak A, Fortuna M, Hryniewicz W. Usefulness of CHROMagar Candida medium, biochemical methods—API ID32C and VITEK 2 compact and two MALDI-TOF MS Systems for Candida spp. identification. Pol J Microbiol. 2016;65(1):111–4.
Kim T-H, Kweon OJ, Kim HR, Lee M-K. Identification of uncommon Candida species using commercial identification systems. J Microbiol Biotechnol. 2016;26(12):2206–13.
Lee HS, Shin JH, Choi MJ, Won EJ, Kee SJ, Kim SH, et al. Comparison of the Bruker Biotyper and VITEK MS matrix-assisted laser desorption/ionization time-of-flight mass spectrometry systems using a formic acid extraction method to identify common and uncommon yeast isolates. Ann Lab Med. 2017;37(3):223–30.
Porte L, García P, Braun S, Ulloa MT, Lafourcade M, Montaña A, et al. Head-to-head comparison of microflex LT and Vitek MS systems for routine identification of microorganisms by MALDI-TOF mass spectrometry in Chile. PLoS One. 2017;12(5):e0177929.
Mancini N, De Carolis E, Infurnari L, Vella A, Clementi N, Vaccaro L, et al. Comparative evaluation of the Bruker Biotyper and Vitek MS matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry systems for identification of yeasts of medical importance. J Clin Microbiol. 2013;51(7):2453–7.
Chen JHK, Yam W-C, Ngan AHY, Fung AMY, Woo W-L, Yan M-K, et al. Advantages of using matrix-assisted laser desorption ionization-time of flight mass spectrometry as a rapid diagnostic tool for identification of yeasts and mycobacteria in the clinical microbiological laboratory. J Clin Microbiol. 2013;51(12):3981–7.
Hamprecht A, Christ S, Oestreicher T, Plum G, Kempf VAJ, Göttig S. Performance of two MALDI-TOF MS systems for the identification of yeasts isolated from bloodstream infections and cerebrospinal fluids using a time-saving direct transfer protocol. Med Microbiol Immunol (Berl). 2014;203(2):93–9.
Jamal WY, Ahmad S, Khan ZU, Rotimi VO. Comparative evaluation of two matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) systems for the identification of clinically significant yeasts. Int J Infect Dis IJID Off Publ Int Soc Infect Dis. 2014;26:167–70.
Chao Q-T, Lee T-F, Teng S-H, Peng L-Y, Chen P-H, Teng L-J, et al. Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS One. 2014;9(10):e109376.
Deak E, Charlton CL, Bobenchik AM, Miller SA, Pollett S, McHardy IH, et al. Comparison of the Vitek MS and Bruker microflex LT MALDI-TOF MS platforms for routine identification of commonly isolated bacteria and yeast in the clinical microbiology laboratory. Diagn Microbiol Infect Dis. 2015;81(1):27–33.
Pence MA, McElvania TeKippe E, Wallace MA, Burnham CA. Comparison and optimization of two MALDI-TOF MS platforms for the identification of medically relevant yeast species. Eur J Clin Microbiol Infect Dis Off Publ Eur Soc Clin Microbiol. 2014;33(10):1703–12.
Bader O, Weig M, Taverne-Ghadwal L, Lugert R, Gross U, Kuhns M. Improved clinical laboratory identification of human pathogenic yeasts by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Microbiol Infect Off Publ Eur Soc Clin Microbiol Infect Dis. 2011;17(9):1359–65.
Hou X, Xiao M, Chen SC, Wang H, Yu S-Y, Fan X, et al. Identification and antifungal susceptibility profiles of Candida nivariensis and Candida bracarensis in a multi-center Chinese collection of yeasts. Front Microbiol. 2017;8:5.
Haas M, Grenouillet F, Loubersac S, Ariza B, Pepin-Puget L, Alvarez-Moreno CA, et al. Identification of cryptic Candida species by MALDI-TOF mass spectrometry, not all MALDI-TOF systems are the same: focus on the C. parapsilosis species complex. Diagn Microbiol Infect Dis. 2016;86(4):385–6.
Gouriet F, Ghiab F, Couderc C, Bittar F, Tissot Dupont H, Flaudrops C, et al. Evaluation of a new extraction protocol for yeast identification by mass spectrometry. J Microbiol Methods. 2016;129:61–5.
Denis J, Machouart M, Morio F, Sabou M, Kauffmann-LaCroix C, Contet-Audonneau N, et al. Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identifying clinical Malassezia isolates. J Clin Microbiol. 2017;55(1):90–6.
de Almeida JN, Sztajnbok J, da Silva AR, Vieira VA, Galastri AL, Bissoli L, et al. Rapid identification of moulds and arthroconidial yeasts from positive blood cultures by MALDI-TOF mass spectrometry. Med Mycol. 2016;54(8):885–9.
Vlek A, Kolecka A, Khayhan K, Theelen B, Groenewald M, Boel E, et al. Interlaboratory comparison of sample preparation methods, database expansions, and cutoff values for identification of yeasts by matrix-assisted laser desorption ionization-time of flight mass spectrometry using a yeast test panel. J Clin Microbiol. 2014;52(8):3023–9.
Kolecka A, Khayhan K, Groenewald M, Theelen B, Arabatzis M, Velegraki A, et al. Identification of medically relevant species of arthroconidial yeasts by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2013;51(8):2491–500.
De Carolis E, Vella A, Vaccaro L, Torelli R, Posteraro P, Ricciardi W, et al. Development and validation of an in-house database for matrix-assisted laser desorption ionization-time of flight mass spectrometry-based yeast identification using a fast protein extraction procedure. J Clin Microbiol. 2014;52(5):1453–8.
McTaggart LR, Lei E, Richardson SE, Hoang L, Fothergill A, Zhang SX. Rapid identification of Cryptococcus neoformans and Cryptococcus gattii by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol. 2011;49(8):3050–3.
Pinto A, Halliday C, Zahra M, van Hal S, Olma T, Maszewska K, et al. Matrix-assisted laser desorption ionization-time of flight mass spectrometry identification of yeasts is contingent on robust reference spectra. PLoS One. 2011;6(10):e25712.
Acknowledgments
We thank Nadia Bueno for the writing assistance and Ruben Barrios for the technical support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no competing interests.
Human and Animal Rights and Informed Consent
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
This article is part of the Topical Collection on Clinical Mycology Lab Issues
Rights and permissions
About this article
Cite this article
Taverna, C.G., Mazza, M., Refojo, N. et al. A Comprehensive Analysis of MALDI-TOF MS and Ribosomal DNA Sequencing for Identification of Clinical Yeasts. Curr Fungal Infect Rep 11, 184–189 (2017). https://doi.org/10.1007/s12281-017-0297-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12281-017-0297-2