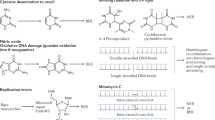
Abstract
Reactive oxygen species induce DNA strand breaks and DNA oxidation. DNA oxidation leads to DNA mismatches, resulting in mutations in the genome if not properly repaired. Homologous recombination (HR) and non-homologous end-joining (NHEJ) are required for DNA strand breaks, whereas the base excision repair system mainly repairs oxidized DNAs, such as 8-oxoguanine and thymine glycol, by cleaving the glycosidic bond, inserting correct nucleotides, and sealing the gap. Our previous studies revealed that the Rad53-Bdr1 pathway mainly controls DNA strand breaks through the regulation of HR- and NHEJ-related genes. However, the functional roles of genes involved in the base excision repair system remain elusive in Cryptococcus neoformans. In the present study, we identified OGG1 and NTG1 genes in the base excision repair system of C. neoformans, which are involved in DNA oxidation repair. The expression of OGG1 was induced in a Hog1-dependent manner under oxidative stress. On the other hand, the expression of NTG1 was strongly induced by DNA damage stress in a Rad53-independent manner. We demonstrated that the deletion of NTG1, but not OGG1, resulted in elevated susceptibility to DNA damage agents and oxidative stress inducers. Notably, the ntg1Δ mutant showed growth defects upon antifungal drug treatment. Although deletion of OGG1 or NTG1 did not increase mutation rates, the mutation profile of each ogg1Δ and ntg1Δ mutant was different from that of the wild-type strain. Taken together, we found that DNA N-glycosylase Ntg1 is required for oxidative DNA damage stress and antifungal drug resistance in C. neoformans.




Similar content being viewed by others
Data availability
Data will be made available on request.
References
Alseth, I., Eide, L., Pirovano, M., Rognes, T., Seeberg, E., & Bjørås, M. (1999). The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Molecular and Cellular Biology, 19, 3779–3787.
Angelé-Martínez, C., Goodman, C., & Brumaghim, J. (2014). Metal-mediated DNA damage and cell death: Mechanisms, detection methods, and cellular consequences. Metallomics, 6, 1358–1381.
Bahn, Y. S., Hicks, J. K., Giles, S. S., Cox, G. M., & Heitman, J. (2004). Adenylyl cyclase-associated protein Aca1 regulates virulence and differentiation of Cryptococcus neoformans via the cyclic AMP-protein kinase A cascade. Eukaryotic Cell, 3, 1476–1491.
Bahn, Y. S., Kojima, K., Cox, G. M., & Heitman, J. (2005). Specialization of the HOG pathway and its impact on differentiation and virulence of Cryptococcus neoformans. Molecular Biology of the Cell, 16, 2285–2300.
Bauer, N. C., Corbett, A. H., & Doetsch, P. W. (2015). The current state of eukaryotic DNA base damage and repair. Nucleic Acids Research, 43, 10083–10101.
Bohr, V. A. (2002). Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radical Biology & Medicine, 32, 804–812.
Boyce, K. J., Wang, Y., Verma, S., Shakya, V. P. S., Xue, C., & Idnurm, A. (2017). Mismatch repair of DNA replication errors contributes to microevolution in the pathogenic fungus Cryptococcus neoformans. mBio, 8, e00595-17.
Choudhary, S., Mundodi, V., Smith, A. D., & Kadosh, D. (2023). Genome-wide translational response of Candida albicans to fluconazole treatment. Microbiology Spectrum, 11, e0257223.
Dbouk, N. H., Covington, M. B., Nguyen, K., & Chandrasekaran, S. (2019). Increase of reactive oxygen species contributes to growth inhibition by fluconazole in Cryptococcus neoformans. BMC Microbiology, 19, 243.
Fan, Y., & Lin, X. (2018). Multiple applications of a transient CRISPR-CAS9 coupled with electroporation (TRACE) system in the Cryptococcus neoformans species complex. Genetics, 208, 1357–1372.
Fukui, M., Choi, H. J., & Zhu, B. T. (2012). Rapid generation of mitochondrial superoxide induces mitochondrion-dependent but caspase-independent cell death in hippocampal neuronal cells that morphologically resembles necroptosis. Toxicology and Applied Pharmacology, 262, 156–166.
Furtado, C., Kunrath-Lima, M., Rajão, M. A., Mendes, I. C., de Moura, M. B., Campos, P. C., Macedo, A. M., Franco, G. R., Pena, S. D., Teixeira, S. M., et al. (2012). Functional characterization of 8-oxoguanine DNA glycosylase of Trypanosoma cruzi. PLoS ONE, 7, e42484.
Giles, S. S., Stajich, J. E., Nichols, C., Gerrald, Q. D., Alspaugh, J. A., Dietrich, F., & Perfect, J. R. (2006). The Cryptococcus neoformans catalase gene family and its role in antioxidant defense. Eukaryotic Cell, 5, 1447–1459.
Griffiths, L. M., Swartzlander, D., Meadows, K. L., Wilkinson, K. D., Corbett, A. H., & Doetsch, P. W. (2009). Dynamic compartmentalization of base excision repair proteins in response to nuclear and mitochondrial oxidative stress. Molecular and Cellular Biology, 29, 794–807.
Hanna, B. M. F., Michel, M., Helleday, T., & Mortusewicz, O. (2021). NEIL1 and NEIL2 are recruited as potential backup for OGG1 upon OGG1 depletion or inhibition by TH5487. International Journal of Molecular Sciences, 22, 4542.
Hoot, S. J., Zheng, X., Potenski, C. J., White, T. C., & Klein, H. L. (2011). The role of Candida albicans homologous recombination factors Rad54 and Rdh54 in DNA damage sensitivity. BMC Microbiology, 11, 214.
Hu, J., de Souza-Pinto, N. C., Haraguchi, K., Hogue, B. A., Jaruga, P., Greenberg, M. M., Dizdaroglu, M., & Bohr, V. A. (2005). Repair of formamidopyrimidines in DNA involves different glycosylases: Role of the OGG1, NTH1, and NEIL1 enzymes. The Journal of Biological Chemistry, 280, 40544–40551.
Iyer, K. R., Revie, N. M., Fu, C., Robbins, N., & Cowen, L. E. (2021). Treatment strategies for cryptococcal infection: Challenges, advances and future outlook. Nature Reviews Microbiology, 19, 454–466.
Iyer, R. R., Pluciennik, A., Burdett, V., & Modrich, P. L. (2006). DNA mismatch repair: Functions and mechanisms. Chemical Reviews, 106, 302–323.
Izawa, S., Inoue, Y., & Kimura, A. (1996). Importance of catalase in the adaptive response to hydrogen peroxide: Analysis of acatalasaemic Saccharomyces cerevisiae. The Biochemical Journal, 320, 61–67.
Jin, J. H., Lee, K. T., Hong, J., Lee, D., Jang, E. H., Kim, J. Y., Lee, Y., Lee, S. H., So, Y. S., Jung, K. W., et al. (2020). Genome-wide functional analysis of phosphatases in the pathogenic fungus Cryptococcus neoformans. Nature Communications, 11, 4212.
Jung, K. W., Jung, J. H., & Park, H. Y. (2021). Functional roles of homologous recombination and non-homologous end joining in DNA damage response and microevolution in Cryptococcus neoformans. Journal of Fungi, 7, 566.
Jung, K. W., Kwon, S., Jung, J. H., & Bahn, Y. S. (2022). Essential roles of ribonucleotide reductases under DNA damage and replication stresses in Cryptococcus neoformans. Microbiology Spectrum, 10, e0104422.
Jung, K. W., Lee, K. T., So, Y. S., & Bahn, Y. S. (2018). Genetic manipulation of Cryptococcus neoformans. Current Protocols in Microbiology, 50, e59.
Jung, K. W., Lee, Y., Huh, E. Y., Lee, S. C., Lim, S., & Bahn, Y. S. (2019). Rad53- and Chk1-dependent DNA damage response pathways cooperatively promote fungal pathogenesis and modulate antifungal drug susceptibility. mBio, 10, e01726-18.
Jung, K. W., Yang, D. H., Kim, M. K., Seo, H. S., Lim, S., & Bahn, Y. S. (2016). Unraveling fungal radiation resistance regulatory networks through the genome-wide transcriptome and genetic analyses of Cryptococcus neoformans. mBio, 7, e10483-16.
Jung, K. W., Yang, D. H., Maeng, S., Lee, K. T., So, Y. S., Hong, J., Choi, J., Byun, H. J., Kim, H., Bang, S., et al. (2015). Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nature Communications, 6, 6757.
Kim, M. S., Kim, S. Y., Yoon, J. K., Lee, Y. W., & Bahn, Y. S. (2009). An efficient gene-disruption method in Cryptococcus neoformans by double-joint PCR with NAT-split markers. Biochemical Biophysical Research Communications, 390, 983–988.
Ko, Y. J., Yu, Y. M., Kim, G. B., Lee, G. W., Maeng, P. J., Kim, S. S., Floyd, A., Heitman, J., & Bahn, Y. S. (2009). Remodeling of global transcription patterns of Cryptococcus neoformans genes mediated by the stress-activated HOG signaling pathways. Eukaryotic Cell, 8, 1197–1217.
Kouchakdjian, M., Bodepudi, V., Shibutani, S., Eisenberg, M., Johnson, F., Grollman, A. P., & Patel, D. J. (1991). NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry, 30, 1403–1412.
Kozmin, S., Slezak, G., Reynaud-Angelin, A., Elie, C., de Rycke, Y., Boiteux, S., & Sage, E. (2005). UVA radiation is highly mutagenic in cells that are unable to repair 7,8-dihydro-8-oxoguanine in Saccharomyces cerevisiae. Proceedings of the National Academy of Sciences of the USA, 102, 13538–13543.
Krokan, H. E., Standal, R., & Slupphaug, G. (1997). DNA glycosylases in the base excision repair of DNA. Biochemical Journal, 325, 1–16.
Legrand, M., Chan, C. L., Jauert, P. A., & Kirkpatrick, D. T. (2008). Analysis of base excision and nucleotide excision repair in Candida albicans. Microbiology, 154, 2446–2456.
Lia, D., Reyes, A., de Melo Campos, J. T. A., Piolot, T., Baijer, J., Radicella, J. P., & Campalans, A. (2018). Mitochondrial maintenance under oxidative stress depends on mitochondrially localised α-OGG1. Journal of Cell Sciences, 131, jcs213538.
Liu, H., & Zhang, J. (2019). Yeast spontaneous mutation rate and spectrum vary with environment. Current Biology, 29, 1584–1591.
Melnikova, V. O., & Ananthaswamy, H. N. (2005). Cellular and molecular events leading to the development of skin cancer. Mutation Research, 571, 91–106.
Melo, R. G., Leitao, A. C., & Pádula, M. (2004). Role of OGG1 and NTG2 in the repair of oxidative DNA damage and mutagenesis induced by hydrogen peroxide in Saccharomyces cerevisiae: Relationships with transition metals iron and copper. Yeast, 21, 991–1003.
Oliveira, R. K. M., Hurtado, F. A., Gomes, P. H., Puglia, L. L., Ferreira, F. F., Ranjan, K., Albuquerque, P., Pocas-Fonseca, M. J., Silva-Pereira, I., & Fernandes, L. (2021). Base excision repair AP-endonucleases-like genes modulate DNA damage response and virulence of the human pathogen Cryptococcus neoformans. Journal of Fungi, 7, 133.
Peng, C. A., Gaertner, A. A. E., Henriquez, S. A., Fang, D., Colon-Reyes, R. J., Brumaghim, J. L., & Kozubowski, L. (2018). Fluconazole induces ROS in Cryptococcus neoformans and contributes to DNA damage in vitro. PLoS ONE, 13, e0208471.
Posteraro, B., Sanguinetti, M., Sanglard, D., La Sorda, M., Boccia, S., Romano, L., Morace, G., & Fadda, G. (2003). Identification and characterization of a Cryptococcus neoformans ATP binding cassette (ABC) transporter-encoding gene, CnAFR1, involved in the resistance to fluconazole. Molecular Microbiology, 47, 357–371.
Rodero, L., Mellado, E., Rodriguez, A. C., Salve, A., Guelfand, L., Cahn, P., Cuenca-Estrella, M., Davel, G., & Rodriguez-Tudela, J. L. (2003). G484S amino acid substitution in lanosterol 14-α demethylase (ERG11) is related to fluconazole resistance in a recurrent Cryptococcus neoformans clinical isolate. Antimicrobial Agents and Chemotherapy, 47, 3653–3656.
Scott, A. D., Neishabury, M., Jones, D. H., Reed, S. H., Boiteux, S., & Waters, R. (1999). Spontaneous mutation, oxidative DNA damage, and the roles of base and nucleotide excision repair in the yeast Saccharomyces cerevisiae. Yeast, 15, 205–218.
Sentürker, S., Auffret van der Kemp, P., You, H. J., Doetsch, P. W., Dizdaroglu, M., & Boiteux, S. (1998). Substrate specificities of the Ntg1 and Ntg2 proteins of Saccharomyces cerevisiae for oxidized DNA bases are not identical. Nucleic Acids Research, 26, 5270–5276.
Singh, K. K., Sigala, B., Sikder, H. A., & Schwimmer, C. (2001). Inactivation of Saccharomyces cerevisiae OGG1 DNA repair gene leads to an increased frequency of mitochondrial mutants. Nucleic Acids Research, 29, 1381–1388.
Sionov, E., Lee, H., Chang, Y. C., & Kwon-Chung, K. J. (2010). Cryptococcus neoformans overcomes stress of azole drugs by formation of disomy in specific multiple chromosomes. PLoS Pathogens, 6, e1000848.
So, Y. S., Maeng, S., Yang, D. H., Kim, H., Lee, K. T., Yu, S. R., Tenor, J. L., Giri, V. K., Toffaletti, D. L., Arras, S., et al. (2019). Regulatory mechanism of the atypical AP-1-like transcription factor Yap1 in Cryptococcus neoformans. mSphere, 4, e1000848.
Steenken, S. (1997). Electron transfer in DNA? Competition by ultra-fast proton transfer? Journal of Biological Chemistry, 378, 1293–1297.
Thomas, D., Scot, A. D., Barbey, R., Padula, M., & Boiteux, S. (1997). Inactivation of OGG1 increases the incidence of G·C→T·A transversions in Saccharomyces cerevisiae: Evidence for endogenous oxidative damage to DNA in eukaryotic cells. Molecular and General Genetics, 254, 171–178.
Upadhya, R., Kim, H., Jung, K. W., Park, G., Lam, W., Lodge, J. K., & Bahn, Y. S. (2013). Sulphiredoxin plays peroxiredoxin-dependent and -independent roles via the HOG signalling pathway in Cryptococcus neoformans and contributes to fungal virulence. Molecular Microbiology, 90, 630–648.
Valentine, J. S., Wertz, D. L., Lyons, T. J., Liou, L. L., Goto, J. J., & Gralla, E. B. (1998). The dark side of dioxygen biochemistry. Current Opinion in Chemical Biology, 2, 253–262.
Vermes, A., Guchelaar, H. J., & Dankert, J. (2000). Flucytosine: A review of its pharmacology, clinical indications, pharmacokinetics, toxicity and drug interactions. Journal of Antimicrobial Chemotherapy, 46, 171–179.
Wang, K., Maayah, M., Sweasy, J. B., & Alnajjar, K. S. (2021). The role of cysteines in the structure and function of OGG1. Journal of Biological Chemistry, 296, 100093.
Wang, P. (2018). Two distinct approaches for CRISPR-CAS9-mediated gene editing in Cryptococcus neoformans and related species. mSphere, 3, e00208-18.
Acknowledgements
This work was supported by the KAERI Institutional R&D program (523430-23).
Funding
Korea Atomic Energy Research Institute R&D program, 523430-23, Kwang-Woo Jung.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there are no competing interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jung, KW., Kwon, S., Jung, JH. et al. Functional Characterization of DNA N-Glycosylase Ogg1 and Ntg1 in DNA Damage Stress of Cryptococcus neoformans. J Microbiol. 61, 981–992 (2023). https://doi.org/10.1007/s12275-023-00092-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-023-00092-y