Abstract
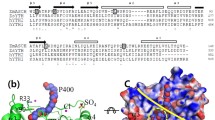
The SbcCD complex is an essential component of the DNA double-strand break (DSB) repair system in bacteria. The bacterial SbcCD complex recognizes and cleaves the DNA ends in DSBs by ATP-dependent endo- and exonuclease activities as an early step of the DNA repair process. SbcD consists of nuclease, capping, and helix-loop-helix domains. Here, we present the crystal structure of a SbcD fragment from Staphylococcus aureus, which contained nuclease and capping domains, at a resolution of 2.9 Å. This structure shows a dimeric assembly similar to that of the corresponding domains of SbcD from Escherichia coli. The S. aureus SbcD fragment exhibited endonuclease activities on supercoiled DNA and exonuclease activity on linear and nicked DNA. This study contributes to the understanding of the molecular basis for how bacteria can resist sterilizing treatment, causing DNA damage.
Similar content being viewed by others
References
Afonine, P.V., Mustyakimov, M., Grosse-Kunstleve, R.W., Moriarty, N.W., Langan, P., and Adams, P.D. 2010. Joint X-ray and neutron refinement with phenix.refine. Acta Crystallogr. D Biol. Crystallogr. 66, 1153–1163.
Bentchikou, E., Servant, P., Coste, G., and Sommer, S. 2007. Additive effects of SbcCD and PolX deficiencies in the in vivo repair of DNA double-strand breaks in Deinococcus radiodurans. J. Bacteriol. 189, 4784–4790.
Bergdoll, M.S. 1991. Staphylococcus aureus. J. Assoc. Off. Anal. Chem. 74, 706–710.
Bhakdi, S. and Tranum-Jensen, J. 1991. Alpha-toxin of Staphylococcus aureus. Microbiol. Rev. 55, 733–751.
Connelly, J.C., de Leau, E.S., and Leach, D.R. 1999. DNA cleavage and degradation by the SbcCD protein complex from Escherichia coli. Nucleic Acids Res. 27, 1039–1046.
Connelly, J.C., de Leau, E.S., and Leach, D.R. 2003. Nucleolytic processing of a protein-bound DNA end by the E. coli SbcCD (MR) complex. DNA Repair 2, 795–807.
Connelly, J.C., Kirkham, L.A., and Leach, D.R. 1998. The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc. Natl. Acad. Sci. USA 95, 7969–7974.
Diebold-Durand, M.L., Lee, H., Ruiz Avila, L.B., Noh, H., Shin, H.C., Im, H., Bock, F.P., Bürmann, F., Durand, A., Basfeld, A., et al. 2017. Structure of full-length SMC and rearrangements required for chromosome organization. Mol. Cell 67, 334–347.
Emsley, P. and Cowtan, K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132.
Hanawalt, P.C., Cooper, P.K., Ganesan, A.K., and Smith, C.A. 1979. DNA repair in bacteria and mammalian cells. Annu. Rev. Biochem. 48, 783–836.
Hohl, M., Kwon, Y., Galván, S.M., Xue, X., Tous, C., Aguilera, A., Sung, P., and Petrini, J.H. 2011. The Rad50 coiled-coil domain is indispensable for Mre11 complex functions. Nat. Struct. Mol. Biol. 18, 1124–1131.
Hopfner, K.P., Craig, L., Moncalian, G., Zinkel, R.A., Usui, T., Owen, B.A., Karcher, A., Henderson, B., Bodmer, J.L., McMurray, C.T., et al. 2002. The Rad50 zinc-hook is a structure joining Mre11 complexes in DNA recombination and repair. Nature 418, 562–566.
Käshammer, L., Saathoff, J.H., Lammens, K., Gut, F., Bartho, J., Alt, A., Kessler, B., and Hopfner, K.P. 2019. Mechanism of DNA end sensing and processing by the Mre11-Rad50 complex. Mol. Cell 76, 382–394.
Lammens, K., Bemeleit, D.J., Möckel, C., Clausing, E., Schele, A., Hartung, S., Schiller, C.B., Lucas, M., Angermüller, C., Soding, J., et al. 2011. The Mre11:Rad50 structure shows an ATP-dependent molecular clamp in DNA double-strand break repair. Cell 145, 54–66.
Lavin, M.F. 2007. ATM and the Mre11 complex combine to recognize and signal DNA double-strand breaks. Oncogene 26, 7749–7758.
Lee, J.H. and Paull, T.T. 2007. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene 26, 7741–7748.
Liu, S., Tian, L., Liu, Y., An, X., Tang, Q., Yan, X., and Liang, D. 2014. Structural basis for DNA recognition and nuclease processing by the Mre11 homologue SbcD in double-strand breaks repair. Acta Crystallogr. D Biol. Crystallogr. 70, 299–309.
Lowy, F.D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339, 520–532.
Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326.
Park, Y.B., Hohl, M., Padjasek, M., Jeong, E., Jin, K.S., Kręźel, A., Petrini, J.H., and Cho, Y. 2017. Eukaryotic Rad50 functions as a rod-shaped dimer. Nat. Struct. Mol. Biol. 24, 248–257.
Saathoff, J.H., Käshammer, L., Lammens, K., Byrne, R.T., and Hopfner, K.P. 2018. The bacterial Mre11-Rad50 homolog SbcCD cleaves opposing strands of DNA by two chemically distinct nuclease reactions. Nucleic Acids Res. 46, 11303–11314.
Sancar, A., Lindsey-Boltz, L.A., Ünsal-Kaçmaz, K., and Linn, S. 2004. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85.
Storvik, K.A.M. and Foster, P.L. 2011. The SMC-like protein complex SbcCD enhances DNA polymerase IV-dependent spontaneous mutation in Escherichia coli. J. Bacteriol. 193, 660–669.
Stracker, T.H., Theunissen, J.W., Morales, M., and Petrini, J.H. 2004. The Mre11 complex and the metabolism of chromosome breaks: the importance of communicating and holding things together. DNA Repair 3, 845–854.
Wigley, D.B. 2013. Bacterial DNA repair: recent insights into the mechanism of RecBCD, AddAB and AdnAB. Nat. Rev. Microbiol. 11, 9–13.
Williams, R.S., Moncalian, G., Williams, J.S., Yamada, Y., Limbo, O., Shin, D.S., Groocock, L.M., Cahill, D., Hitomi, C., Guenther, G., et al. 2008. Mre11 dimers coordinate DNA end bridging and nuclease processing in double-strand-break repair. Cell 135, 97–109.
Williams, R.S., Williams, J.S., and Tainer, J.A. 2007. Mre11-Rad50-Nbs1 is a keystone complex connecting DNA repair machinery, double-strand break signaling, and the chromatin template. Biochem. Cell Biol. 85, 509–520.
Winn, M.D., Ballard, C.C., Cowtan, K.D., Dodson, E.J., Emsley, P., Evans, P.R., Keegan, R.M., Krissinel, E.B., Leslie, A.G., McCoy, A., et al. 2011. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242.
Wyman, C. and Kanaar, R. 2006. DNA double-strand break repair: all’s well that ends well. Annu. Rev. Genet. 40, 363–383.
Acknowledgements
This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Ministry of Science & ICT (grant NRF-2017M3A9F6029755, NRF-2019M3E5D6063871). We made use of beamlines 11C at the Pohang Accelerator Laboratory (Pohang, Republic of Korea) and the MALS facility at the Korea Basic Science Institute (Ochang, Republic of Korea).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
We have no conflicts of interest to report.
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Lee, J., Jo, I., Ahn, J. et al. Crystal structure of the nuclease and capping domain of SbcD from Staphylococcus aureus. J Microbiol. 59, 584–589 (2021). https://doi.org/10.1007/s12275-021-1012-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-021-1012-0