Abstract
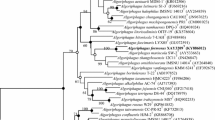
A novel, Gram-stain-negative marine bacterium, designated GH2-6T, was isolated from a rhizosphere mudflat of a halophyte (Carex scabrifolia) collected in Gangwha Island, the Republic of Korea. The cells of the organism were strictly aerobic, oxidase- and catalase-positive, non-flagellated rods. Growth occurred at 20–45°C, pH 5–10, and 0.5–9 (w/v) NaCl. The requirement of Na+ for growth (0.5–3%) was observed. The major respiratory quinone was Q-10. The major polar lipids were phosphatidylcholine, phosphatidylethanolamine, phosphatidylglycerol, an aminolipid and a glycolipid. The predominant fatty acids were C18:1ω7c, C18:0, C16:0, C19:0 cyclo ω8c, C18:1ω7c 11-methyl and summed feature 2 (C14:0 3-OH and/or C16:1 iso I). The genome size was 4.45 Mb and the G+C content of the genomic DNA was 61.9 mol%. Phylogenetic analyses based on 16S rRNA gene sequences revealed that strain GH2-6T belonged to genus Martelella and formed a tight cluster with M. radicis BM5-7T and M. endophytica YC6887T. Levels of 16S rRNA gene sequence similarity between the novel isolate and members of the genus were 99.3–95.5%, but strain GH2-6T possessed an extended loop (49 nucleotides in length) between positions 187 and 213 of the 16S rRNA gene sequence (E. coli numbering). DDH values in vitro between the novel isolate and the closest relatives were 23.2±12.8–46.3±5.2%. On the basis of polyphasic data presented in this study, the type strain GH2-6T (= KACC 19403T = KCTC 62125T = NBRC 113212T) represents a novel species of the genus Martelella for which the name Martelella lutilitoris sp. nov. is proposed.
Similar content being viewed by others
References
Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Bibi, F., Chung, E.J., Khan, A., Jeon, C.O., and Chung, Y.R. 2013. Martelella endophytica sp. nov., an antifungal bacterium associated with a halophyte. Int. J. Syst. Evol. Microbiol. 63, 2914–2919.
Brosius, J., Palmer, M.L., Kennedy, P.J., and Noller, H.F. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75, 4801–4805.
Chen, C.X., Zhang, X.Y., Liu, C., Yu, Y., Liu, A., Li, G.W., Li, H., Chen, X.L., Chen, B., Zhou, B.C., et al. 2013. Pseudorhodobacter antarcticus sp. nov., isolated from Antarctic intertidal sandy sediment, and emended description of the genus Pseudorhodobacter Uchino et al. 2002 emend. Jung et al. 2012. Int. J. Syst. Evol. Microbiol. 63, 849–854.
Chung, E.J., Hwang, J.M., Kim, K.H., Jeon, C.O., and Chung, Y.R. 2016. Martelella suaedae sp. nov. and Martelella limonii sp. nov., isolated from the root of halophytes. Int. J. Syst. Evol. Microbiol. 66, 3917–3922.
Ezaki, T., Hashimoto, Y., and Yabuuchi, E. 1989. Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int. J. Syst. Bacteriol. 39, 224–229.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Felsenstein, J. 2002. PHYLIP (phylogenetic inference package), version 3.6a. Department of Genone Sciences, University of Washington, Seattle, USA.
Fitch, W.M. 1971. Towards defining the course of evolution: minimum change for a specific tree topology. System. Zool. 20, 406–416.
Goris, J., Konstantinidis, K.T., Klappenbach, J.A., Coenye, T., Vandamme, P., and Tiedje, J.M. 2007. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91.
Jukes, T.H. and Cantor, C.R. 1969. Evolution of protein molecules, pp. 21–132. In Munro, H.N. (ed.), Mammalian Protein Metabolism. Academic Press, New York, USA.
Jung, Y.T., Park, S., Lee, J.S., and Yoon, J.H. 2017. Pseudorhodobacter ponti sp. nov., isolated from seawater. Int. J. Syst. Evol. Microbiol. 67, 1855–1860.
Kroppenstedt, R.M. 1985. Fatty acid and menaquinone analysis of actinomycetes and related organisms, pp. 173–199. In Goodfellow, M. and Minnikin, D.E. (eds.). Chemical Methods in Bacterial Systematics, Academic Press, London, UK.
Lane, D.J. 1991. 16S/23S rRNA Sequencing, pp. 115–175. In Stackebrandt, E. and Goodfellow, M. (eds.). Nucleic Acid Techniques in Bacterial Systematics, John Wiley and Sons, London, UK.
Lee, S.D. 2019. Martelella caricis sp. nov., isolated from a rhizosphere mudflat. Int. J. Syst. Evol. Microbiol. 69, 266–270.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Minnikin, D.E., Patel, P.V., Alshamaony, L., and Goodfellow, M. 1977. Polar lipid composition in the classification of Nocardia and related bacteria. Int. J. Syst. Bacteriol. 27, 104–117.
Richter, M. and Rosselló-Móra, R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131.
Rivas, R., Sánchez-Márquez, S., Mateos, P.F., Martínez-Molina, E., and Velázquez, E. 2005. Martelella mediterranea gen. nov., sp. nov., a novel α-proteobacterium isolated from a subterranean saline lake. Int. J. Syst. Evol. Microbiol. 55, 955–959.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Stackebrandt, E. and Goebel, B.M. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44, 846–849.
Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. 1997. The Clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24, 4876–4882.
Uchino, Y., Hamada, T., and Yokota, A. 2002. Proposal of Pseudorhodobacter ferrugineus gen. nov., comb. nov., for a non-photosynthetic marine bacterium, Agrobacterium ferrugineum, related to the genus Rhodobacter. J. Gen. Appl. Microbiol. 48, 309–320.
Wayne, L.G., Brenner, D.J., Colwell, R.R., Grimont, P.A.D., Kandler, O., Krichevsky, M.I., Moore, L.H., Moore, W.E.C., Murray, R.G.E., Stackebrandt, E., et al. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Bacteriol. 37, 463–464.
Zhang, D.C. and Margesin, R. 2014. Martelella radicis sp. nov. and Martelella mangrovi sp. nov., isolated from mangrove sediment. Int. J. Syst. Evol. Microbiol. 64, 3104–3108.
Acknowledgements
This research was carried out by the project for the survey and excavation of Korean indigenous species of the National Institute of Biological Resources (NIBR) under the Ministry of Environment, Korea, and through the partial support of the National Research Foundation of Korea (no. 2019015605). We thank D. B. Jeon for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kim, YJ., Lee, S.D. Martelella lutilitoris sp. nov., isolated from a tidal mudflat. J Microbiol. 57, 976–981 (2019). https://doi.org/10.1007/s12275-019-9259-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-9259-4