Abstract
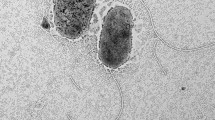
A Gram-stain-negative, rod-shaped, obligately aerobic, chemoheterotrophic bacterium which is motile by means of a single polar flagellum, designated SAORIC-263T, was isolated from deep seawater of the Pacific Ocean. Phylogenetic analyses based on 16S rRNA gene sequences and genomebased phylogeny revealed that strain SAORIC-263T belonged to the genus Sulfitobacter and shared 96.1–99.9% 16S rRNA gene sequence similarities with Sulfitobacter species. Wholegenome sequencing of strain SAORIC-263T revealed a genome size of 3.9Mbp and DNA G+C content of 61.3 mol%. The SAORIC-263T genome shared an average nucleotide identity and digital DNA-DNA hybridization of 79.1–88.5% and 18.9–35.0%, respectively, with other Sulfitobacter genomes. The SAORIC-263T genome contained the genes related to benzoate degradation, which are frequently found in deep-sea metagenome. The strain contained summed feature 8 (C18:1ω7c), C18:1ω7c 11-methyl, and C16:0 as the predominant cellular fatty acids as well as ubiquinone-10 (Q-10) as the major respiratory quinone. The major polar lipids of the strain were phosphatidylethanolamine, phosphatidylglycerol, diphosphatidylglycerol, phosphatidylcholine, and aminolipid. On the basis of taxonomic data obtained in this study, it is suggested that strain SAORIC-263T represents a novel species of the genus Sulfitobacter, for which the name Sulfitobacter profundi sp. nov. is proposed. The type strain is SAORIC-263T (= KACC 21183T = NBRC 113428T).
Similar content being viewed by others
References
Choo, Y.J., Lee, K., Song, J., and Cho, J.C. 2007. Puniceicoccus vermicola gen. nov., sp. nov., a novel marine bacterium, and description of Puniceicoccaceae fam. nov., Puniceicoccales ord. nov., Opitutaceae fam. nov., Opitutales ord. nov. and Opitutae classis nov. in the phylum ‘Verrucomicrobia’. Int. J. Syst. Evol. Microbiol. 57, 532–537.
Collins, M.D., Shah, H.N., and Minnikin, D.E. 1980. A note on the separation of natural mixtures of bacterial menaquinones using reverse phase thin–layer chromatography. J. Appl. Bacteriol. 48, 277–282.
Curson, A.R., Rogers, R., Todd, J.D., Brearley, C.A., and Johnston, A.W. 2008. Molecular genetic analysis of a dimethylsulfoniopropionate lyase that liberates the climate–changing gas dimethylsulfide in several marine alpha–proteobacteria and Rhodobacter sphaeroides. Environ. Microbiol. 10, 757–767.
Eloe, E.A., Malfatti, F., Gutierrez, J., Hardy, K., Schmidt, W.E., Pogliano, K., Pogliano, J., Azam, F., and Bartlett, D.H. 2011. Isolation and characterization of the first psychropiezophilic alphaproteobacterium. Appl. Environ. Microbiol. 77, 8145–8153.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Felsenstein, J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791.
Fukui, Y., Abe, M., Kobayashi, M., and Satomi, M. 2015. Sulfitobacter pacificus sp. nov., isolated from the red alga Pyropia yezoensis. Antonie van Leeuwenhoek 107, 1155–1163.
Fukui, Y., Abe, M., Kobayashi, M., Shimada, Y., Saito, H., Oikawa, H., Yano, Y., and Satomi, M. 2014. Sulfitobacter porphyrae sp. nov., isolated from the red alga Porphyra yezoensis. Int. J. Syst. Evol. Microbiol. 64, 438–443.
Goris, J., Konstantinidis, K.T., Klappenbach, J.A., Coenye, T., Vandamme, P., and Tiedje, J.M. 2007. DNA–DNA hybridization values and their relationship to whole–genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91.
Honda, M.C., Wakita, M., Matsumoto, K., Fujiki, T., Siswanto, E., Sasaoka, K., Kawakami, H., Mino, Y., Sukigara, C., and Kitamura, M. 2017. Comparison of carbon cycle between the western Pacific subarctic and subtropical time–series stations: highlights of the K2S1 project. J. Oceanogr. 73, 647–667.
Ivanova, E.P., Gorshkova, N.M., Sawabe, T., Zhukova, N.V., Hayashi, K., Kurilenko, V.V., Alexeeva, Y., Buljan, V., Nicolau, D.V., Mikhailov, V.V., et al. 2004. Sulfitobacter delicatus sp. nov. and Sulfitobacter dubius sp. nov., respectively from a starfish (Stellaster equestris) and sea grass (Zostera marina). Int. J. Syst. Evol. Microbiol. 54, 475–480.
Kanehisa, M., Sato, Y., and Morishima, K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome Sequences. J. Mol. Biol. 428, 726–731.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Kumari, P., Bhattacharjee, S., Poddar, A., and Das, S.K. 2016. Sulfitobacter faviae sp. nov., isolated from the coral Favia veroni. Int. J. Syst. Evol. Microbiol. 66, 3786–3792.
Kwak, M.J., Lee, J.S., Lee, K.C., Kim, K.K., Eom, M.K., Kim, B.K., and Kim, J.F. 2014. Sulfitobacter geojensis sp. nov., Sulfitobacter noctilucae sp. nov., and Sulfitobacter noctilucicola sp. nov., isolated from coastal seawater. Int. J. Syst. Evol. Microbiol. 64, 3760–3767.
Labrenz, M., Tindall, B.J., Lawson, P.A., Collins, M.D., Schumann, P., and Hirsch, P. 2000. Staleya guttiformis gen. nov., sp. nov. and Sulfitobacter brevis sp. nov., α–3–Proteobacteria from hypersaline, heliothermal and meromictic Antarctic Ekho Lake. Int. J. Syst. Evol. Microbiol. 50, 303–313.
Liu, Y., Lai, Q., and Shao, Z. 2017. Proposal for transfer of Oceanibulbus indolifex Wagner–Döbler et al. 2004 to the genus Sulfitobacter as Sulfitobacter indolifex comb. nov. Int. J. Syst. Evol. Microbiol. 67, 2328–2331.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., et al. 2004. ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Markowitz, V.M., Mavromatis, K., Ivanova, N.N., Chen, I.M., Chu, K., and Kyrpides, N.C. 2009. IMG ER: a system for microbial ge nome annotation expert review and curation. Bioinformatics 25, 2271–2278.
Martín–Cuadrado, A.B., López–García, P., Alba, J.C., Moreira, D., Monticelli, L., Strittmatter, A., Gottschalk, G., and Rodríguez–Valera, F. 2007. Metagenomics of the deep Mediterranean, a warm bathypelagic habitat. PLoS One 2, e914.
Meier–Kolthoff, J.P., Auch, A.F., Klenk, H.P., and Goker, M. 2013. Genome sequence–based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Na, S.I., Kim, Y.O., Yoon, S.H., Ha, S.M., Baek, I., and Chun, J. 2018. UBCG: Up–to–date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 56, 281–285.
Park, A.Y., Teeravet, S., Pheng, S., Lee, J.R., Kim, S.G., and Suwannachart, C. 2018. Sulfitobacter aestuarii sp. nov., a marine bacterium isolated from a tidal flat of the Yellow Sea. Int. J. Syst. Evol. Microbiol. 68, 1771–1775.
Pruesse, E., Peplies, J., and Glockner, F.O. 2012. SINA: accurate high–throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28, 1823–1829.
Pukall, R., Buntefuss, D., Fruhling, A., Rohde, M., Kroppenstedt, R.M., Burghardt, J., Lebaron, P., Bernard, L., and Stackebrandt, E. 1999. Sulfitobacter mediterraneus sp. nov., a new sulfite–oxidizing member of the alpha–Proteobacteria. Int. J. Syst. Evol. Microbiol. 49, 513–519.
Richter, M. and Rossello–Mora, R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131.
Rzhetsky, A. and Nei, M. 1993. Theoretical foundation of the minimum–evolution method of phylogenetic inference. Mol. Biol. Evol. 10, 1073–1095.
Saitou, N. and Nei, M. 1987. The neighbor–joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. MIDI Inc., Newark, DE, USA.
Song, J., Kang, I., Joung, Y., Yoshizawa, S., Kaneko, R., Oshima, K., Hattori, M., Hamasaki, K., Kogure, K., Kim, S., et al. 2019. Genome analysis of Rubritalea profundi SAORIC–165T, the first deep–sea verrucomicrobial isolate, from the northwestern Pacific Ocean. J. Microbiol. In press.
Sorokin, D.Y. 1995. Sulfitobacter pontiacus gen. nov., sp. nov. a new heterotrophic bacterium from the Black Sea, specialized on sulfite oxidation. Mikrobiologiya 64, 295–295.
Stothard, P. and Wishart D.S. 2005. Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539.
Weisburg, W.G., Barns, S.M., Pelletier, D.A., and Lane, D.J. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703.
Yoon, S.H., Ha, S.M., Kwon, S., Lim, J., Kim, Y., Seo, H., and Chun, J. 2017a. Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole–genome assemblies. Int. J. Syst. Evol. Microbiol. 67, 1613–1617.
Yoon, S.H., Ha, S.M., Lim, J., Kwon, S., and Chun, J. 2017b. A largescale evaluation of algorithms to calculate average nucleotide identity. Antonie van Leeuwenhoek 110, 1281–1286.
Yoon, J.H., Kang, S.J., and Oh, T.K. 2007. Sulfitobacter marinus sp. nov., isolated from seawater of the East Sea in Korea. Int. J. Syst. Evol. Microbiol. 57, 302–305.
Acknowledgements
We are grateful to the officer and crew of R/V Mirai (Japan Agency for Marine-Earth Science and Technology [JAMSTEC]) for their assistance and support with sample collection. We would also like to thank Dr. Kazuhiro Kogure for his support with the isolation of deep-sea bactrial strains.
This study was supported by Inha University Research Grant.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Song, J., Jang, HJ., Joung, Y. et al. Sulfitobacter profundi sp. nov., isolated from deep seawater. J Microbiol. 57, 661–667 (2019). https://doi.org/10.1007/s12275-019-9150-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-9150-3