Abstract
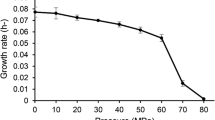
A Gram-positive, aerobic, rod-shaped, spore-forming bacterium, designated YLB-03T, with peritrichous flagella was isolated from deep-sea sediment of the Yap Trench at a depth of 4435 m. The bacterium was found to be catalase-positive but oxidase-negative. Growth of this bacterium was observed at 15–50°C (optimum 37°C), pH 5–10.5 (optimum 7), 0–5% NaCl (optimum 1%, w/v) and 0.1–50 MPa (optimum 0.1 MPa). Phylogenetic analysis based on 16S rRNA gene sequences showed that strain YLB-03T was a member of the genus Lysinibacillus. Strain YLB-03T was closely related to Lysinibacillus sinduriensis BLB-1T and Lysinibacillus chungkukjangi 2RL3-2T (98.4%), Lysinibacillus halotolerans LAM-612T (98.0%), Lysinibacillus telephonicus KT735049T (97.5%), Lysinibacillus endophyticus C9T (97.5%), Lysinibacillus composti NCCP-36T and Lysinibacillus massiliensis 4400831T (97.3%). The ANI and the GGDC DNA-DNA hybridization estimate values between strain YLB-03T and closely related type strains were 73.7–76.3% and 34.7–38.7%, respectively. The principal fatty acids were anteiso-C15:0 and iso-C15:0. The G+C content of the chromosomal DNA was 39.6 mol%. The respiratory quinone was determined to be MK-7. The diagnostic amino acids in the cell wall peptidoglycan contained Lys-Asp (type A4α) and the cell-wall sugars were glucose and xylose. The polar lipids included diphosphatidylglycerol, phosphatidylglycerol, phosphatidylethanolamine, and an unidentified phospholipid. The combined genotypic and phenotypic data showed that strain YLB-03T represents a novel species within the genus Lysinibacillus, for which the name Lysinibacillus yapensis sp. nov. is proposed, with the type strain YLB-03T (= MCCC 1A12698T = JCM 32871T).
Similar content being viewed by others
References
Ahmed, I., Yokota, A., Yamazoe, A., and Fujiwara, T. 2007. Proposal of Lysinibacillus boronitolerans gen. nov. sp. nov., and transfer of Bacillus fusiformis to Lysinibacillus fusiformis comb. nov. and Bacillus sphaericus to Lysinibacillus sphaericus comb. nov. Int. J. Syst. Evol. Microbiol. 57, 1117–1125.
Auch, A.F., Klenk, H.P., and Göker, M. 2010a. Standard operating procedure for calculating genome-to-genome distances based on high-scoring segment pairs. Stand. Genomic Sci. 2, 142–148.
Auch, A.F., von Jan, M., Klenk, H.P., and Göker, M. 2010b. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2, 117–134.
Azmatunnisa, M., Rahul, K., Lakshmi, K.V.N.S., Sasikala, Ch., and Ramana, Ch.V. 2015. Lysinibacillus acetophenoni sp. nov., a solvent-tolerant bacterium isolated from acetophenone. Int. J. Syst. Evol. Microbiol. 65, 1741–1748.
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A.A., Dvorkin, M., Kulikov, A.S., Lesin, V.M., Nikolenko, S.I., Pham, S., Prjibelski, A.D., et al. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477.
Begum, M.A., Rahul, K., Sasikala, C., and Ramana, C.V. 2016. Lysinibacillus xyleni sp. nov., isolated from a bottle of xylene. Arch. Microbiol. 198, 325–332.
Cheng, M., Zhang, H., Zhang, J., Hu, G., Zhang, J., He, J., and Huang, X. 2015. Lysinibacillus fluoroglycofenilyticus sp. nov., a bacterium isolated from fluoroglycofen contaminated soil. Antonie van Leeuwenhoek 107, 157–164.
Coorevits, A., Dinsdale, A.E., Heyrman, J., Schumann, P., Van Landschoot, A., Logan, N.A., and De Vos, P. 2012. Lysinibacillus macroides sp. nov., nom. rev. Int. J. Syst. Evol. Microbiol. 62, 1121–1127.
Duan, Y.Q., He, S.T., Li, Q.Q., Wang, M.F., Wang, W.Y., Zhe, W., Cao, Y.H., Mo, M.H., Zhai, Y.L., and Li, W.J. 2013. Lysinibacillus tabacifolii sp. nov., a novel endophytic bacterium isolated from Nicotiana tabacum leaves. J. Microbiol. 51, 289–294.
Fang, J., Zhang, L., and Bazylinski, D.A. 2010. Deep-sea piezosphere and piezophiles: geomicrobiology and biogeochemistry. Trends Microbiol. 18, 413–422.
Felsenstein, J. 1981. Evolutionary trees from DNA sequences: a maximum likelihood approach. J. Mol. Evol. 17, 368–376.
Glazunova, O.O., Raoult, D., and Roux, V. 2006. Bacillus massiliensis sp. nov., isolated from cerebrospinal fluid. Int. J. Syst. Evol. Microbiol. 56, 1485–1488.
Goris, J., Konstantinidis, K.T., Klappenbach, J.A., Coenye, T., Vandamme, P., and Tiedje, J.M. 2007. DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91.
Hayat, R., Ahmed, I., Paek, J., Sin, Y., Ehsan, M., Iqbal, M., Yokota, A., and Chang, Y.H. 2014. Lysinibacillus composti sp. nov., isolated from compost. Ann. Microbiol. 64, 1081–1088.
Jamieson, A.J., Fujii, T., Mayor, D.J., Solan, M., and Priede, I.G. 2010. Hadal trenches: the ecology of the deepest places on Earth. Trends Ecol. Evol. 25, 190–197.
Jung, M.Y., Kim, J.S., Paek, W.K., Styrak, I., Park, LS., Sin, Y., Paek, J., Park, K.A., Kim, H., Kim, H.L., et al. 2012. Description of Lysinibacillus sinduriensis sp. nov., and transfer of Bacillus massiliensis and Bacillus odysseyi to the genus Lysinibacillus as Lysinibacillus massiliensis comb. nov. and Lysinibacillus odysseyi comb. nov. with emended description of the genus Lysinibacillus. Int. J. Syst. Evol. Microbiol. 62, 2347–2355.
Kämpfer, P., Martin, K., and Glaeser, S.P. 2013. Lysinibacillus contaminans sp. nov., isolated from surface water. Int. J. Syst. Evol. Microbiol 63, 3148–3153.
Kim, O.S., Cho, Y.J., Lee, K., Yoon, S.H., Kim, M., Na, H., Park, S.C., Jeon, Y.S., Lee, J.H., Yi, H., et al. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721.
Kim, S.J., Jang, Y.H., Hamada, M., Ahn, J.H., Weon, H.Y., Suzuki, K.I., Whang, K.S., and Kwon, S.W. 2013a. Lysinibacillus chungkukjangi sp. nov., isolated from Chungkukjang, Korean fermented soybean food. J. Microbiol. 51, 400–404.
Kim, J.Y., Park, S.H., Oh, D.C., and Kim, Y.J. 2013b. Lysinibacillus jejuensis sp. nov., isolated from swinery waste. J. Microbiol. 51, 872–876.
Komagata, K. and Suzuki, K.I. 1988. Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol. 19, 161–207.
Kong, D., Wang, Y., Zhao, B., Li, Y., Song, J., Zhai, Y., Zhang, C., Wang, H., Chen, X., Zhao, B., et al. 2014. Lysinibacillus halotolerans sp. nov., isolated from saline-alkaline soil. Int. J. Syst. Evol. Microbiol. 64, 2593–2598.
Kumar, S., Stecher, G., and Tamura, K. 2016. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874.
Lee, C.S., Jung, Y.T., Park, S., Oh, T.K., and Yoon, J.H. 2010. Lysinibacillus xylanilyticus sp. nov., a xylan-degrading bacterium isolated from forest humus. Int. J. Syst. Evol. Microbiol. 60, 281–286.
Liu, H., Song, Y., Chen, F., Zheng, S., and Wang, G. 2013. Lysinibacillus manganicus sp. nov., isolated from manganese mining soil. Int. J. Syst. Evol. Microbiol. 63, 3568–3573.
Meier-Kolthoff, J.P., Auch, A.F., Klenk, H.P., and Göker, M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14, 60.
Minnikin, D.E., O’Donnell, A.G., Goodfellow, M., Alderson, G., Athalye, M., Schaal, A., and Parlett, J.H. 1984. An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J. Microbiol. Methods 2, 233–241.
Miwa, H., Ahmed, I., Yokota, A., and Fujiwara, T. 2009. Lysinibacillus parviboronicapiens sp. nov., a low-boron-containing bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 59, 1427–1432.
Nunoura, T., Takaki, Y., Hirai, M., Shimamura, S., Makabe, A., Koide, O., Kikuchi, T., Miyazaki, J., Koba, K., Yoshida, N., et al. 2016. Hadal biosphere: insight into the microbial ecosystem in the deepest ocean on Earth. Proc. Natl. Acad. Sci. USA 112, 1230–1236.
Ouoba, L.I.I., Vouidibio Mbozo, A.B., Thorsen, L., Anyogu, A., Nielsen, D.S., Kobawila, S.C., and Sutherland, J.P. 2015. Lysinibacillus louembei sp. nov., a spore-forming bacterium isolated from Ntoba Mbodi, alkaline fermented leaves of cassava from the Republic of the Congo. Int. J. Syst. Evol. Microbiol. 65, 4256–4262.
Rahi, P., Kurli, R., Khairnar, M., Jagtap, S., Pansare, A.N., Dastager, S.G., and Shouche, Y.S. 2017. Description of Lysinibacillus telephonicus sp. nov., isolated from the screen of a cellular phone. Int. J. Syst. Evol. Microbiol. 67, 2289–2295.
Ren, Y., Chen, S.Y., Yao, H.Y., and Deng, L.J. 2015. Lysinibacillus cresolivorans sp. nov., an m-cresol-degrading bacterium isolated from coking wastewater treatment aerobic sludge. Int. J. Syst. Evol. Microbiol. 65, 4250–4255.
Richter, M. and Rosselló-Móra, R. 2009. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 106, 19126–19131.
Rzhetsky, A. and Nei, M. 1992. Statistical properties of the ordinary least-squares, generalized least-squares, and minimum-evolution methods of phylogenetic inference. J. Mol. Evol. 35, 367–375.
Saitou, N. and Nei, M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425.
Sasser, M. 1990. Identification of bacteria by gas chromatography of cellular fatty acids. MIDI technical note 101. MIDI, Newark, DE, USA.
Schleifer, K.H. 1985. 5 Analysis of the chemical composition and primary structure of murein. Methods Microbiol. 18, 123–156.
Schleifer, K.H. and Kandler, O. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36, 407–477.
Staneck, J.L. and Roberts, G.D. 1974. Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl. Microbiol. 28, 226–231.
Tindall, B.J. 1990. Lipid composition of Halobacterium lacusprofundi. FEMS Microbiol. Lett. 66, 199–202.
Wang, J., Li, J., Dasgupta, S., Zhang, L., Golovko, M.Y., Golovko, S.A., and Fang, J. 2014. Alterations in membrane phospholipid fatty acids of Gram-positive piezotolerant bacterium Sporosarcina sp. DSK25 in response to growth pressure. Lipids 49, 347–356.
Wayne, L.G., Brenner, D.J., Colwell, R.R., Grimont, P.A.D., Kandler, O., Krichevsky, M.I., Moore, L.H., Moore, W.E.C., Murray, R.G.E., Stackebrandt, E., et al. 1987. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int. J. Syst. Evol. Microbiol. 37, 463–464.
Yang, L.L., Huang, Y., Liu, J., Ma, L., Mo, M.H., Li, W.J., and Yang, F.X. 2012. Lysinibacillus mangiferahumi sp. nov., a new bacterium producing nematicidal volatiles. Antonie van Leeuwenhoek 102, 53–59.
Yayanos, A.A. 2001. Deep-sea piezophilic bacteria. Methods Microbiol. 30, 615–637.
Yu, J., Guan, X., Liu, C., Xiang, W., Yu, Z., Liu, X., and Wang, G. 2016. Lysinibacillus endophyticus sp. nov., an indole-3-acetic acid producing endophytic bacterium isolated from corn root (Zea mays cv. Xinken-5). Antonie van Leeuwenhoek 109, 1337–1344.
Yu, L., Lai, Q., Yi, Z., Zhang, L., Huang, Y., Gu, L., and Tang, X. 2013. Microbacterium sediminis sp. nov., a psychrotolerant, thermotolerant, halotolerant, and alkalitolerant actinomycete isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 63, 25–30.
Zhang, Z., Wu, Y., and Zhang, X.H. 2017. Cultivation of microbes from the deep-sea environments. Deep Sea Res. Part II Top. Stud. Oceanogr. 155, 34–43.
Zhao, F., Feng, Y., Chen, R., Zhang, J., and Lin, X. 2015. Lysinibacillus alkaliphilus sp. nov., an extremely alkaliphilic bacterium, and emended description of genus Lysinibacillus. Int. J. Syst. Evol. Microbiol. 65, 2426–2431.
Zhu, C., Sun, G., Chen, X., Guo, J., and Xu, M. 2014. Lysinibacillus varians sp. nov., an endospore-forming bacterium with a filament-to-rod cell cycle. Int. J. Syst. Evol. Microbiol. 64, 3644–3649.
Acknowledgements
This work was supported by the National Basic Research Program of China (973 Program) (No.2015CB755901), COMRA Project of China (DY135-B2-16), Fujian Key Science and Technology Program (No.2018N0017), Xiamen Ocean Economic Innovation and Development Demonstration Project (No.16PZP001SF16), Scientific Research Foundation of Third Institute of Oceanography, SOA. (No.2016002, 2017002), Xiamen Science and Technology Program (No. 3502Z2018-2029), and Ministry of Natural Resources supporting project of Marine Biodiversity of Areas National Jurisdiction, No. HC180701. We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Supplemental material for this article may be found at http://www.springerlink.com/content/120956.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Yu, L., Tang, X., Wei, S. et al. Isolation and characterization of a novel piezotolerant bacterium Lysinibacillus yapensis sp. nov., from deep-sea sediment of the Yap Trench, Pacific Ocean. J Microbiol. 57, 562–568 (2019). https://doi.org/10.1007/s12275-019-8709-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12275-019-8709-3