Abstract
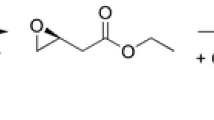
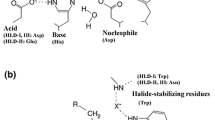
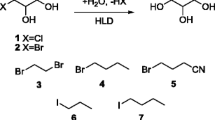
Halohydrin dehalogenases (HHDHs) are biotechnologically relevant enzymes that can be applied as biocatalysts for the selective synthesis of various β-substituted alcohols. Despite that fact, only very few HHDHs are currently available. In an attempt to identify novel ones, database mining of publicly available sequence databases was performed using HHDH-specific sequence information. As a result, 19 novel HHDHs were obtained all exhibiting true HHDH activity.
Similar content being viewed by others
Literatur
Iqbal HA, Feng Z, Brady SF (2012) Biocatalysts and small molecule products from metagenomic studies. Curr Opin Chem Biol 16:109–116
Höhne M, Schätzle S, Jochens H et al. (2010) Rational assignment of key motifs for function guides in silico enzyme identification. Nature Chem Biol 6:807–813
Schallmey M, Floor RJ, Szymanski W et al. (2012) Halohydrin dehalogenases. In: Carreira EM, Yamamoto H (Hrsg) Comprehensive chirality. Elsevier, Amsterdam, 143–155
van Hylkama Vlieg JET, Tang L, Lutje Spelberg JH et al. (2001) Halohydrin dehalogenases are structurally and mechanistically related to short-chain dehydrogenases/reductases. J Bacteriol 183:5058–5066
Yu F, Nakamura T, Mizunashi W et al. (1994) Cloning of two halohydrin hydrogen-halide-lyase genes of Corynebacterium sp. strain N-1074 and structural comparison of the genes and gene products. Biosci Biotech Biochem 58:605–612
Persson B, Kallberg Y (2013) Classification and nomenclature of the superfamily of short-chain dehydrogenases/reductases (SDRs). Chem Biol Interact 202:111–115
de Jong RM, Tiesinga JJW, Rozeboom HJ et al. (2003) Structure and mechanism of a bacterial haloalcohol dehalogenase: a new variation of the short-chain dehydrogenase/reductase fold without an NAD(P)H binding site. EMBO J 22:4933–4944
Schallmey M, Floor RJ, Hauer B et al. (2013) Biocatalytic and structural properties of a highly engineered halohydrin dehalogenase. ChemBioChem 14:870–881
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Schallmey, A., Schallmey, M. & Wardenga, R. Identifikation neuartiger Halohydrin-Dehalogenasen. Biospektrum 19, 816–817 (2013). https://doi.org/10.1007/s12268-013-0394-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12268-013-0394-x